3L4J
 
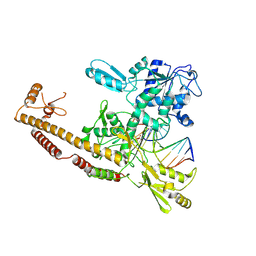 | | Topoisomerase II-DNA cleavage complex, apo | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
3HOG
 
 | |
3HM3
 
 | |
3K6T
 
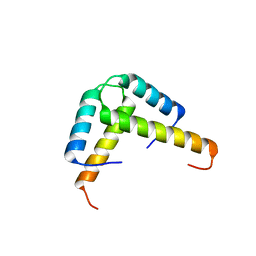 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans at 2.04 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3JBW
 
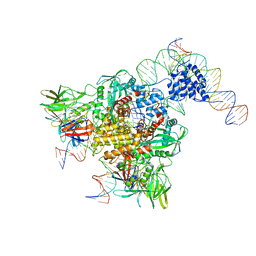 | | Cryo-electron microscopy structure of RAG Paired Complex (with NBD, no symmetry) | | Descriptor: | 12-RSS signal end forward strand, 5'-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', Nicked 12-RSS intermediate reverse strand, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3KBL
 
 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3KBE
 
 | | Metal-free C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
3KSQ
 
 | | Discovery of C-Imidazole Azaheptapyridine FPT Inhibitors | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyltransferase, CAAX box, ... | | Authors: | Zhu, H.Y, Cooper, A.B, Njoroge, G, Kirschmeier, P, Strickland, C, Hruza, A, Girijavallabhan, V.M. | | Deposit date: | 2009-11-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of C-imidazole azaheptapyridine FPT inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L4K
 
 | | Topoisomerase II-DNA cleavage complex, metal-bound | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
3IOZ
 
 | |
3HOL
 
 | |
3HOE
 
 | |
3KLY
 
 | |
3KLZ
 
 | |
3KN8
 
 | |
4OTB
 
 | |
1SNK
 
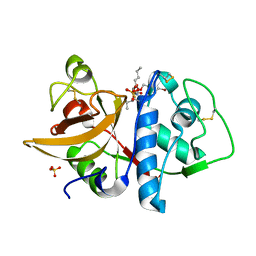 | | Cathepsin K complexed with carbamate derivatized norleucine aldehyde | | Descriptor: | Cathepsin K, N2-({[(4-BROMOPHENYL)METHYL]OXY}CARBONYL)-N1-[(1S)-1-FORMYLPENTYL]-L-LEUCINAMIDE, SULFATE ION | | Authors: | Boros, E.E, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Thompson, J.B, Willard Jr, D.H, Wright, L.L. | | Deposit date: | 2004-03-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploration of the P(2)-P(3) SAR of aldehyde cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SI0
 
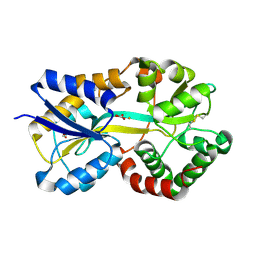 | | Crystal Structure of Mannheimia haemolytica Ferric iron-Binding Protein A in a closed conformation | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, Snell, G, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens.
J.Bacteriol., 186, 2004
|
|
1T3F
 
 | | THREE DIMENSIONAL STRUCTURE OF A HUMANIZED ANTI-IFN-GAMMA FAB (HuZAF) IN P21 21 21 SPACE GROUP | | Descriptor: | Huzaf antibody heavy chain, Huzaf antibody light chain | | Authors: | Bourne, P.C, Terzyan, S.S, Cloud, G, Landolfi, N.F, Vasquez, M, Edmundson, A.B. | | Deposit date: | 2004-04-26 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of a humanized anti-IFN-gamma Fab (HuZAF) in two crystal forms.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T04
 
 | | Three dimensional structure of a humanized anti-IFN-Gamma Fab in C2 space group | | Descriptor: | Huzaf Antibody Heavy Chain, Huzaf Antibody Light Chain | | Authors: | Bourne, P.C, Terzyan, S.S, Cloud, G, Landolfi, N.F, Vasquez, M, Edmundson, A.B. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of a humanized anti-IFN-gamma Fab (HuZAF) in two crystal forms.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T66
 
 | | The structure of FAB with intermediate affinity for fluorescein. | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, immunoglobulin heavy chain, immunoglobulin light chain | | Authors: | Terzyan, S, Ramsland, P.A, Voss Jr, E.W, Herron, J.N, Edmundson, A.B. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional Structures of Idiotypically Related Fabs with Intermediate and High Affinity for Fluorescein.
J.Mol.Biol., 339, 2004
|
|
1UB5
 
 | | Crystal structure of Antibody 19G2 with hapten at 100K | | Descriptor: | 4-(4-STYRYL-PHENYLCARBAMOYL)-BUTYRIC ACID, antibody 19G2, alpha chain, ... | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
1TTK
 
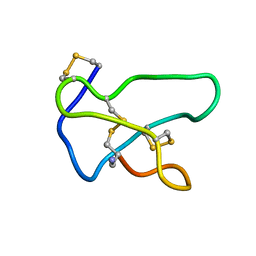 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TT3
 
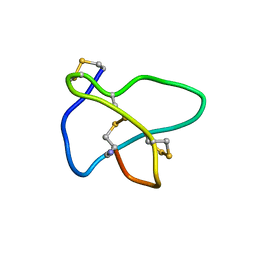 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1U8E
 
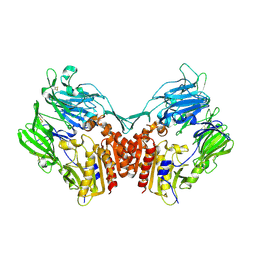 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
