7ENP
 
 | | wild type of O type Foot-and-mouth disease virus | | Descriptor: | VP1 of O type FMDV capsid protein, VP2 of O type FMDV capsid protein, VP3 of O type FMDV capsid protein, ... | | Authors: | Dong, H, Lu, Y. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
5XK9
 
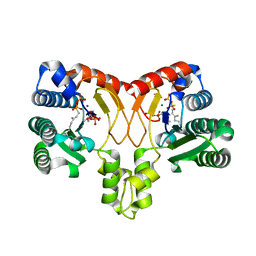 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GSPP and DMAPP | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3BJM
 
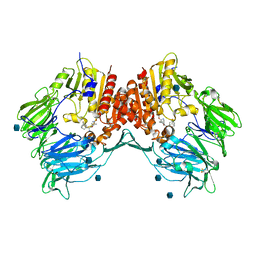 | | Crystal structure of human DPP-IV in complex with (1S,3S, 5S)-2-[(2S)-2-AMINO-2-(3-HYDROXYTRICYCLO[3.3.1.13,7]DEC-1- YL)ACETYL]-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (CAS), (1S,3S,5S)-2-((2S)-2-AMINO-2-(3-HYDROXYADAMANTAN-1- YL)ACETYL)-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (IUPAC), OR BMS-477118 | | Descriptor: | (2~{S})-2-azanyl-1-[(1~{S},3~{S},5~{S})-3-(iminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(5~{R},7~{S})-3-oxidanyl-1-ad amantyl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2007-12-04 | | Release date: | 2008-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Involvement of DPP-IV catalytic residues in enzyme-saxagliptin complex formation.
Protein Sci., 17, 2008
|
|
6JJ3
 
 | | BRD4 in complex with 138A | | Descriptor: | 2-methoxy-N-[2-methyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
5ZG9
 
 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | Authors: | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
6JX4
 
 | |
5Z50
 
 | | Crystal structure of PaCysB regulatory domain | | Descriptor: | Cys regulon transcriptional activator, GLYCEROL, SULFATE ION | | Authors: | Yang, C, Liang, H, Gan, J. | | Deposit date: | 2018-01-15 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa.
Mol.Microbiol., 111, 2019
|
|
3C6D
 
 | | The pseudo-atomic structure of dengue immature virus | | Descriptor: | Polyprotein, prM | | Authors: | Li, L. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | The flavivirus precursor membrane-envelope protein complex: structure and maturation
Science, 319, 2008
|
|
7YIJ
 
 | | Human KCNH5 pore dilation but the non-conducting state | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
3C5X
 
 | | Crystal structure of the precursor membrane protein- envelope protein heterodimer from the dengue 2 virus at low pH | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]alpha-D-altropyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The flavivirus precursor membrane-envelope protein complex: structure and maturation.
Science, 319, 2008
|
|
7DWY
 
 | | S protein of SARS-CoV-2 in the locked conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX6
 
 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 3 (2 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DWX
 
 | | Conformation 1 of S-ACE2-B0AT1 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX3
 
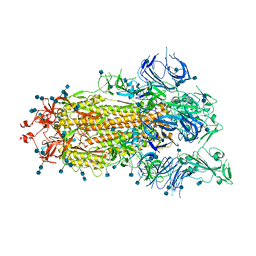 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 1 (1 up RBD and no PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX0
 
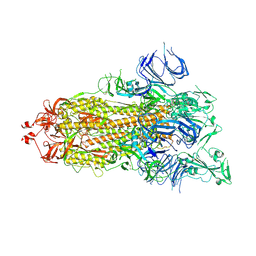 | | Trypsin-digested S protein of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX1
 
 | | S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DWZ
 
 | | S protein of SARS-CoV-2 in the active conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX2
 
 | | Trypsin-digested S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX8
 
 | | Trypsin-digested S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 2 (2 up RBD and 2 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX5
 
 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 2 (1 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX9
 
 | | Trypsin-digested S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 3 (3 up RBD and 2 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX7
 
 | | Trypsin-digested S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 1 (1 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
3AFK
 
 | |
3SWJ
 
 | | Crystal structure of Campylobacter jejuni ChuZ | | Descriptor: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | Authors: | Hu, Y. | | Deposit date: | 2011-07-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
