7DHX
 
 | | Crystal structure of SARS-CoV-2 RBD binding to pangolin ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ZINC ION, ... | | Authors: | Wang, Q.H, Qi, J.X, Wu, L.L. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of pangolin ACE2 engaged by COVID-19 virus
Chin.Sci.Bull., 66, 2021
|
|
7XYO
 
 | | Crystal Structure of a M61 aminopeptidase family member from Myxococcus fulvus | | Descriptor: | Aminopeptidase M61, HEXAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chen, X, Wang, X, Huo, L, Wu, D. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of a Myxobacterial Lanthipeptide with Unique Biosynthetic Features and Anti-inflammatory Activity.
J.Am.Chem.Soc., 145, 2023
|
|
7VNI
 
 | | AHR-ARNT PAS-B heterodimer | | Descriptor: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNA
 
 | | drosophlia AHR PAS-B domain | | Descriptor: | Ahr homolog spineless | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-10 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNH
 
 | |
7W8S
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
 | |
7UFH
 
 | | Integrin alaphIIBbeta3 complex with fradafiban (Mn/Ca) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-22 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99768162 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UDH
 
 | | Integrin alaphIIBbeta3 complex with BMS4-3 | | Descriptor: | (4-{[(5S)-3-(piperidin-4-yl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99998844 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UK9
 
 | | Integrin alaphIIBbeta3 complex with lamifiban (Mn) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.60001969 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UKT
 
 | | Integrin alaphIIBbeta3 complex with BMS4.2 | | Descriptor: | (1-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperidin-4-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36994433 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UCY
 
 | | Integrin alaphIIBbeta3 complex with gantofiban | | Descriptor: | (4-{[(5R)-3-(4-carbamimidoylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 Fab light chain, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34996319 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7U9V
 
 | | Integrin alaphIIBbeta3 complex with BMS4-1 | | Descriptor: | (4-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 light chain, ... | | Authors: | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25492167 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UKO
 
 | | Integrin alaphIIBbeta3 complex with sibrafiban (Mn) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UJK
 
 | | Integrin alaphIIBbeta3 complex with lamifiban | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.43265581 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UDG
 
 | | Integrin alaphIIBbeta3 complex with lotrafiban | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.800069 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UJE
 
 | | Integrin alaphIIBbeta3 complex with UR2922 in Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.499968 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UE0
 
 | | Integrin alaphIIBbeta3 complex with fradafiban | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.741962 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UKP
 
 | | Integrin alaphIIBbeta3 complex with a gantofiban analog | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.80112982 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UBR
 
 | | Integrin alaphIIBbeta3 complex with GR144053 | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04992747 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7UH8
 
 | | Integrin alaphIIBbeta3 complex with roxifiban (Mn/Ca) | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.750024 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7XNN
 
 | | human KCNQ1-CaM-ML277-PIP2 complex in state B | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VM0
 
 | | Crystal structure of YojK from B.subtilis in complex with UDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glycosyl transferase family 1, ... | | Authors: | Hou, X.D, Guo, B.D, Rao, Y.J. | | Deposit date: | 2021-10-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK.
Front Bioeng Biotechnol, 10, 2022
|
|
7V2Y
 
 | | cryo-EM structure of yeast THO complex with Sub2 | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7V2W
 
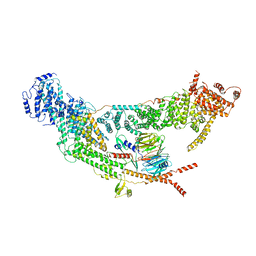 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
