5CB2
 
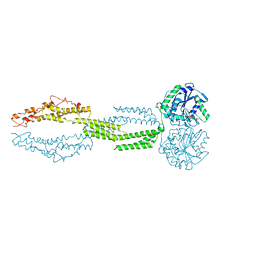 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
2RLI
 
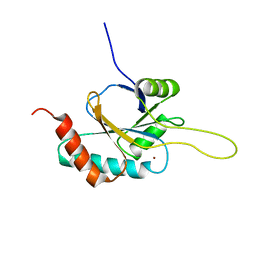 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
3OH6
 
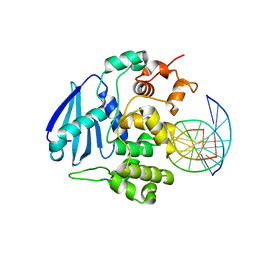 | | AlkA Undamaged DNA Complex: Interrogation of a C:G base pair | | Descriptor: | 5'-D(*GP*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*CP*AP*TP*TP*CP*AP*TP*GP*TP*C)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
3OGD
 
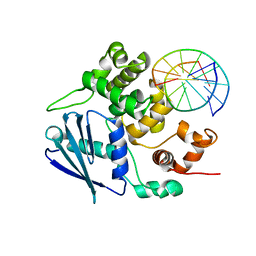 | | AlkA Undamaged DNA Complex: Interrogation of a G*:C base pair | | Descriptor: | 5'-D(*CP*AP*(BRU)P*GP*AP*CP*(BRU)P*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*CP*AP*TP*G)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
1X7A
 
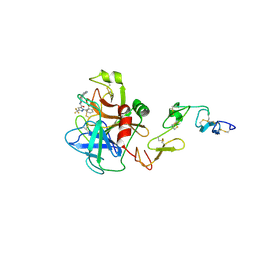 | | Porcine Factor IXa Complexed to 1-{3-[amino(imino)methyl]phenyl}-N-[4-(1H-benzimidazol-1-yl)-2-fluorophenyl]-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide | | Descriptor: | 1-{3-[AMINO(IMINO)METHYL]PHENYL}-N-[4-(1H-BENZIMIDAZOL-1-YL)-2-FLUOROPHENYL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOLE-5-CARBOXAMIDE, Coagulation Factor IX, light chain, ... | | Authors: | Alexander, R.S, Smallwood, A.M, Smallheer, J.M, Wang, J, Wang, S, Nakajima, S, Rossi, K.A, Barbera, F, Burdick, D, Luettgen, J.M. | | Deposit date: | 2004-08-13 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | SAR and factor IXa crystal structure of a dual inhibitor of factors IXa and Xa
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1GYJ
 
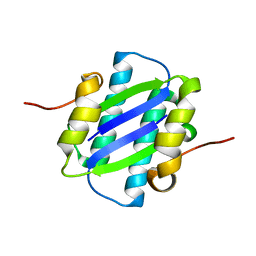 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-23 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1GYY
 
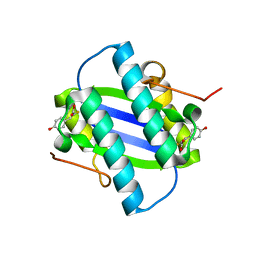 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1GYX
 
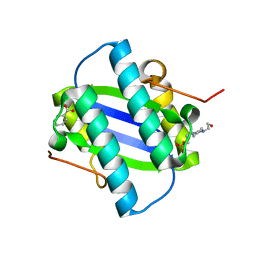 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
3OH9
 
 | | AlkA Undamaged DNA Complex: Interrogation of a T:A base pair | | Descriptor: | 5'-D(*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*TP*TP*CP*AP*TP*GP*T)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
1THN
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1TID
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: Poised for phosphorylation complex with ATP, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
7T3V
 
 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase, spacegroup P22121 after crystals soaked with Zn2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
1L0O
 
 | | Crystal Structure of the Bacillus stearothermophilus Anti-Sigma Factor SpoIIAB with the Sporulation Sigma Factor SigmaF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, MAGNESIUM ION, ... | | Authors: | Campbell, E.A, Masuda, S, Sun, J.L, Muzzin, O, Olson, C.A, Wang, S, Darst, S.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF.
Cell(Cambridge,Mass.), 108, 2002
|
|
1TH8
 
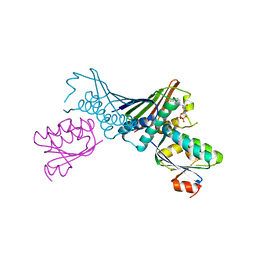 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigian, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
6JJP
 
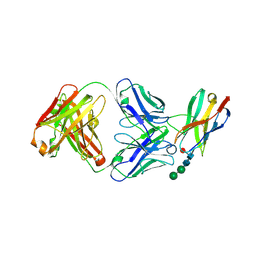 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
7SRV
 
 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase (inactive form), spacegroup P22121 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
5H94
 
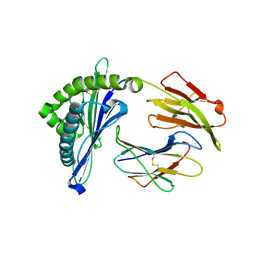 | | Crystal structure of Swine MHC CLASSI for 1.48 angstroms | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nonapeptide from Influenza A virus HA protein | | Authors: | Fan, S, Zhang, N, Wang, S, Wu, Y, Xia, C. | | Deposit date: | 2015-12-25 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Biochemical Analyses of Swine Major Histocompatibility Complex Class I Complexes and Prediction of the Epitope Map of Important Influenza A Virus Strains
J.Virol., 90, 2016
|
|
4ZN7
 
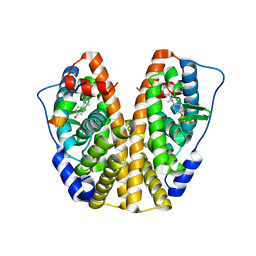 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Diethylstilbestrol | | Descriptor: | DIETHYLSTILBESTROL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNV
 
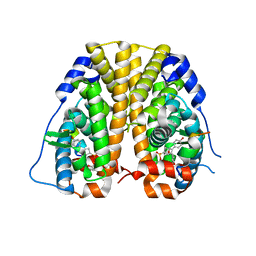 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methoxy-substituted OBHS derivative | | Descriptor: | 2-methoxyphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNS
 
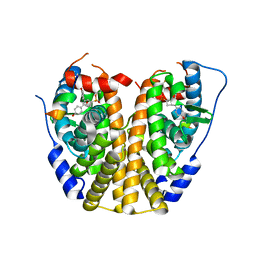 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Fluoro-substituted OBHS derivative | | Descriptor: | 3-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZNU
 
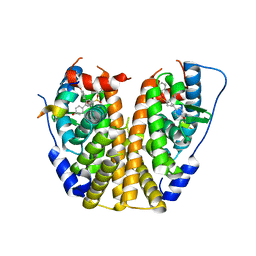 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methyl-substituted OBHS derivative | | Descriptor: | 2-methylphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4JLW
 
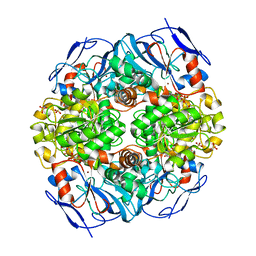 | | Crystal structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | Glutathione-independent formaldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Chen, S, Liao, Y.P, Wang, D.L, Wang, S, Ding, J.F, Wang, Y.M, Cai, L.J, Ran, X.Y, Zhu, H.X. | | Deposit date: | 2013-03-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1TIL
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA:Poised for phosphorylation complex with ATP, crystal form II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
6EE3
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6k) and catalytic zinc ion | | Descriptor: | (1R,2r,3S,5R,7R)-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]tricyclo[3.3.1.1~3,7~]decane-2-carboxamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6EED
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6p) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclohexylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
