3O7M
 
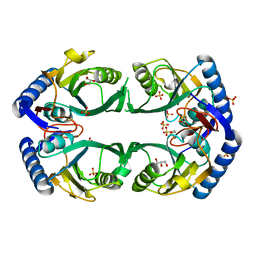 | | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor'
TO BE PUBLISHED
|
|
4EVM
 
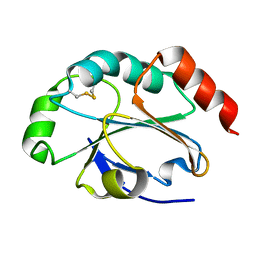 | | 1.5 Angstrom crystal structure of soluble domain of membrane-anchored thioredoxin family protein from Streptococcus pneumoniae strain Canada MDR_19A | | Descriptor: | Thioredoxin family protein | | Authors: | Wawrzak, Z, Stogios, P.J, Minasov, G, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | 1.5 Angstrom crystal structure of soluble domain of membrane-anchored thioredoxin family protein from Streptococcus pneumoniae strain Canada MDR_19A
To be Published
|
|
4EGD
 
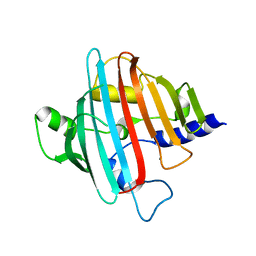 | | 1.85 Angstrom crystal structure of native hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom crystal structure of native hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
3O8Q
 
 | | 1.45 Angstrom Resolution Crystal Structure of Shikimate 5-Dehydrogenase (aroE) from Vibrio cholerae | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of Shikimate 5-Dehydrogenase (aroE) from Vibrio cholerae.
TO BE PUBLISHED
|
|
3OAM
 
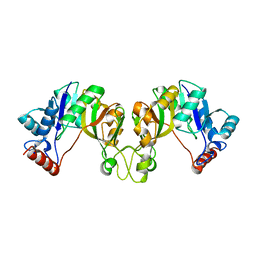 | | Crystal structure of cytidylyltransferase from Vibrio cholerae | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase, SODIUM ION | | Authors: | Hattne, J, Borek, D, Grimshaw, S, Nakka, C, Rostankowski, R, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of cytidylyltransferase from Vibrio cholerae
TO BE PUBLISHED
|
|
3OKF
 
 | | 2.5 Angstrom Resolution Crystal Structure of 3-Dehydroquinate Synthase (aroB) from Vibrio cholerae | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of 3-Dehydroquinate Synthase (aroB) from Vibrio cholerae.
TO BE PUBLISHED
|
|
3P2A
 
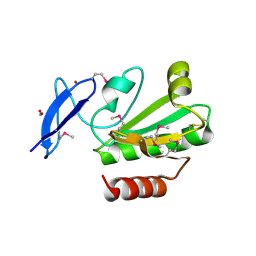 | | Crystal Structure of Thioredoxin 2 from Yersinia pestis | | Descriptor: | FORMIC ACID, Putative thioredoxin-like protein, ZINC ION | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-01 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Crystal Structure of Thioredoxin 2 from Yersinia pestis
TO BE PUBLISHED
|
|
3OT5
 
 | | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes.
TO BE PUBLISHED
|
|
3OJC
 
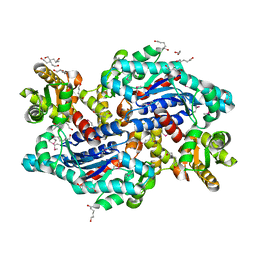 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3P0R
 
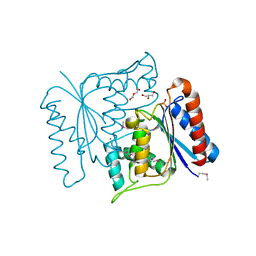 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
3OUU
 
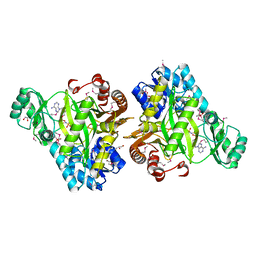 | | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni | | Descriptor: | Biotin carboxylase, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
4EG9
 
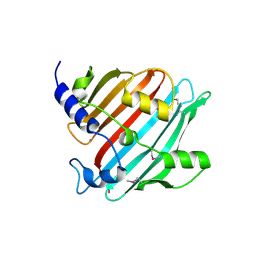 | | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
3P2L
 
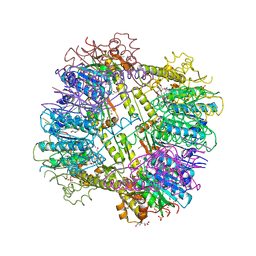 | | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-02 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal Structure of ATP-dependent Clp protease subunit P from Francisella tularensis
To be Published
|
|
3OPQ
 
 | | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure. | | Descriptor: | CHLORIDE ION, FORMIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoribosylaminoimidazole carboxylase with fructose-6-phosphate bound to the central channel of the octameric protein structure.
To be Published
|
|
3KUX
 
 | | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the YPO2259 putative oxidoreductase from Yersinia pestis
To be Published
|
|
3OUZ
 
 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
3P2O
 
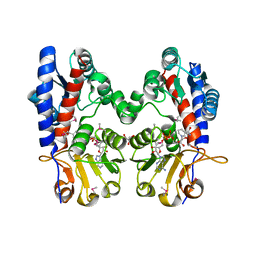 | | Crystal Structure of FolD Bifunctional Protein from Campylobacter jejuni | | Descriptor: | Bifunctional protein folD, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kim, Y, Zhang, R, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Crystal Structure of FolD Bifunctional Protein from
To be Published
|
|
3OXP
 
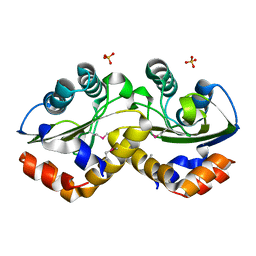 | | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution | | Descriptor: | GLYCEROL, Phosphotransferase enzyme II, A component, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-21 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution
To be Published
|
|
4LES
 
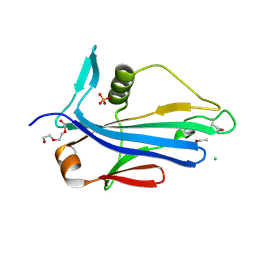 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3M5V
 
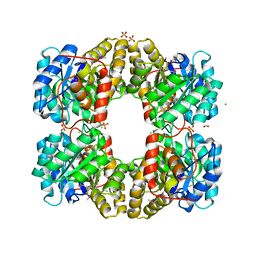 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-28 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni
To be Published
|
|
4NMY
 
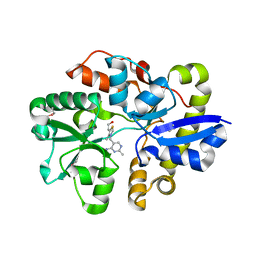 | | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ABC-type transport system, extracellular solute-binding protein | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile
To be Published
|
|
4NOI
 
 | | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase subunit alpha, IODIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4NP6
 
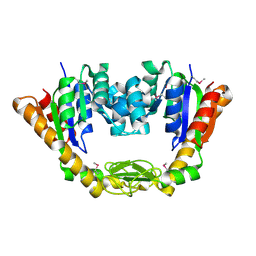 | | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor | | Descriptor: | Adenylate kinase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor
To be Published
|
|
4EQ9
 
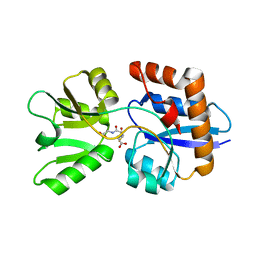 | | 1.4 Angstrom Crystal Structure of ABC Transporter Glutathione-Binding Protein GshT from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Glutathione | | Descriptor: | ABC transporter substrate-binding protein-amino acid transport, GLUTATHIONE | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Light, S.H, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom Crystal Structure of Putative ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Glutathione.
TO BE PUBLISHED
|
|
4NV4
 
 | | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal peptidase I, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.
TO BE PUBLISHED
|
|
