5DA7
 
 | | monomeric PCNA bound to a small protein inhibitor | | Descriptor: | DNA polymerase sliding clamp 1, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Ladner, J.E, Altieri, A.S, Kelman, Z. | | Deposit date: | 2015-08-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp.
Nucleic Acids Res., 44, 2016
|
|
5D6T
 
 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
5DAI
 
 | |
5BXA
 
 | | Structure of PslG from Pseudomonas aeruginosa in complex with mannose | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
3E2Y
 
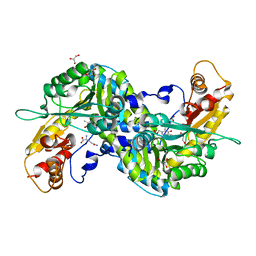 | | Crystal structure of mouse kynurenine aminotransferase III in complex with glutamine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLUTAMINE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, R, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2008-08-06 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Correction for Han et al., "Biochemical and Structural Properties of Mouse Kynurenine Aminotransferase III".
Mol. Cell. Biol., 38, 2018
|
|
3E2Z
 
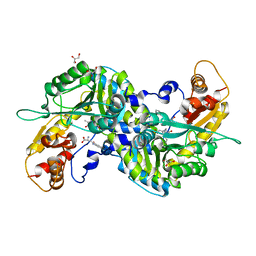 | | Crystal structure of mouse kynurenine aminotransferase III in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, R, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2008-08-06 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Correction for Han et al., "Biochemical and Structural Properties of Mouse Kynurenine Aminotransferase III".
Mol. Cell. Biol., 38, 2018
|
|
3E2F
 
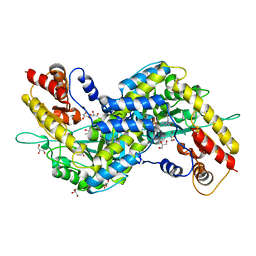 | | Crystal structure of mouse kynurenine aminotransferase III, PLP-bound form | | Descriptor: | GLYCEROL, Kynurenine-oxoglutarate transaminase 3 | | Authors: | Han, Q, Robinson, R, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2008-08-05 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Correction for Han et al., "Biochemical and Structural Properties of Mouse Kynurenine Aminotransferase III".
Mol. Cell. Biol., 38, 2018
|
|
4CAA
 
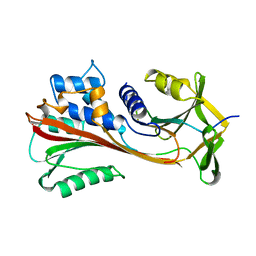 | | CLEAVED ANTICHYMOTRYPSIN T345R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1ZTP
 
 | | X-ray structure of gene product from homo sapiens Hs.433573 | | Descriptor: | Basophilic leukemia expressed protein BLES03 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure at 2.5 A resolution of human basophilic leukemia-expressed protein BLES03.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
3CAA
 
 | | CLEAVED ANTICHYMOTRYPSIN A347R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
441D
 
 | |
1RCW
 
 | |
440D
 
 | |
2H42
 
 | | Crystal structure of PDE5 in complex with sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
2H44
 
 | | Crystal structure of PDE5A1 in complex with icarisid II | | Descriptor: | 5,7-DIHYDROXY-2-(4-METHOXYPHENYL)-8-(3-METHYLBUTYL)-4-OXO-4H-CHROMEN-3-YL 6-DEOXY-ALPHA-L-MANNOPYRANOSIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
4IOY
 
 | |
2QYM
 
 | | crystal structure of unliganded PDE4C2 | | Descriptor: | MAGNESIUM ION, Phosphodiesterase 4C, ZINC ION | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
2QYN
 
 | | Crystal structure of PDE4D2 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
2NNW
 
 | | Alternative conformations of Nop56/58-fibrillarin complex and implication for induced-fit assenly of box C/D RNPs | | Descriptor: | Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein | | Authors: | Oruganti, S, Zhang, Y, Terns, R, Terns, M.P, Li, H. | | Deposit date: | 2006-10-24 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Conformations of the Archaeal Nop56/58-Fibrillarin Complex Imply Flexibility in Box C/D RNPs.
J.Mol.Biol., 371, 2007
|
|
2QYL
 
 | | Crystal structure of PDE4B2B in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, Phosphodiesterase 4B, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
3L2K
 
 | | Structure of phenazine antibiotic biosynthesis protein with substrate | | Descriptor: | EhpF, phenazine-1,6-dicarboxylic acid | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the D-alanylgriseoluteic acid biosynthetic protein EhpF, an atypical member of the ANL superfamily of adenylating enzymes.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3H77
 
 | | Crystal structure of Pseudomonas aeruginosa PqsD in a covalent complex with anthranilate | | Descriptor: | Anthraniloyl-coenzyme A, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
