7WVP
 
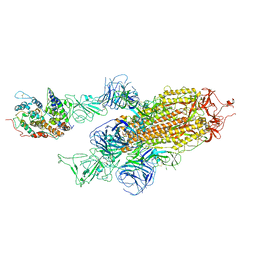 | |
8F22
 
 | |
3EFK
 
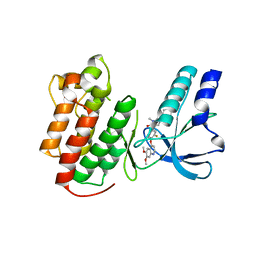 | | Structure of c-Met with pyrimidone inhibitor 50 | | Descriptor: | 5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-2-[(4-fluorophenyl)amino]-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, D'Angelo, N, Whittington, D, Dussault, I. | | Deposit date: | 2008-09-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
7D9O
 
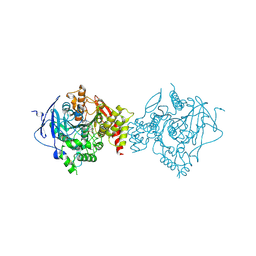 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 2 | | Descriptor: | (2R)-2-[[4-fluoranyl-1-[(4-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
5CSD
 
 | |
5E7X
 
 | | Ligand binding domain 1 of Penicillium marneffei MP1 protein in complex with palmitic acid | | Descriptor: | ACETATE ION, Cell wall antigen, GLYCEROL, ... | | Authors: | Lam, W.H, Zhang, H, Hao, Q. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Talaromyces marneffeiMp1 protein, a novel virulence factor, carries two arachidonic acid-binding domains to suppress inflammatory responses in hosts.
Infect. Immun., 2019
|
|
5ECF
 
 | |
5WJK
 
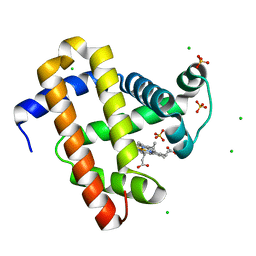 | | 2.0-Angstrom In situ Mylar structure of sperm whale myoglobin (SWMb) at 293 K | | Descriptor: | CHLORIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Broecker, J, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
7D9P
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 12 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(2-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9Q
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 7 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(3-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
5WKT
 
 | | 3.2-Angstrom In situ Mylar structure of bovine opsin at 100 K | | Descriptor: | Rhodopsin, SULFATE ION, Transducin Galpha peptide, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
2LMZ
 
 | |
2Q7M
 
 | | Crystal structure of human FLAP with MK-591 | | Descriptor: | 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2Q7R
 
 | | Crystal structure of human FLAP with an iodinated analog of MK-591 | | Descriptor: | 3-[3-(3,3-DIMETHYLBUTANOYL)-1-(4-IODOBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
7X7D
 
 | | SARS-CoV-2 Delta RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1 | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
7X7E
 
 | | SARS-CoV-2 RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
5XSN
 
 | | The catalytic domain of GdpP with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
7VXA
 
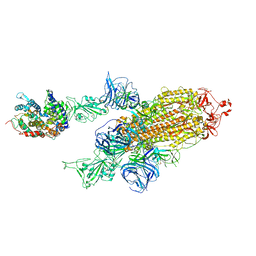 | | SARS-CoV-2 Kappa variant spike protein in complex with ACE2, state C2a | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX9
 
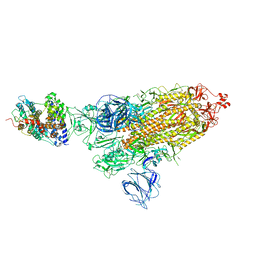 | | SARS-CoV-2 Kappa variant spike protein in complex wth ACE2, state C1 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VX1
 
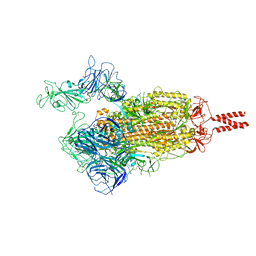 | | SARS-CoV-2 Beta variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXE
 
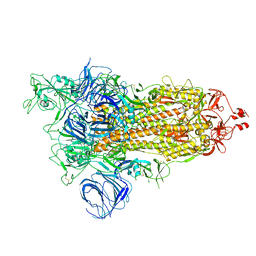 | | SARS-CoV-2 Kappa variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXC
 
 | | SARS-CoV-2 Kappa variant spike protein in C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXM
 
 | |
5XSI
 
 | | The catalytic domain of GdpP | | Descriptor: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSP
 
 | | The catalytic domain of GdpP with 5'-pApA | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
