1N9M
 
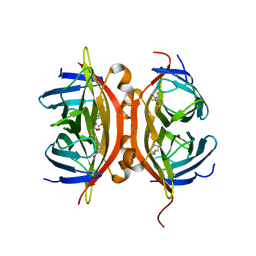 | | Streptavidin Mutant S27A with Biotin at 1.6A Resolution | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-25 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N9Y
 
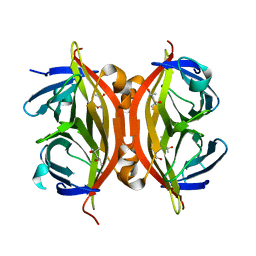 | | Streptavidin Mutant S27A at 1.5A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1DF8
 
 | | S45A MUTANT OF STREPTAVIDIN IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Hyre, D.E, Le Trong, I, Freitag, S, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 1999-11-18 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system.
Protein Sci., 9, 2000
|
|
1N4J
 
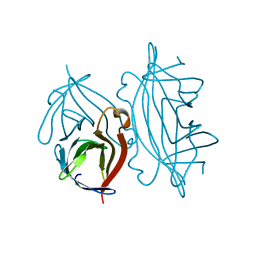 | | STREPTAVIDIN MUTANT N23A AT 2.18A | | Descriptor: | Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-10-31 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NC9
 
 | | STREPTAVIDIN MUTANT Y43A WITH IMINOBIOTIN AT 1.8A RESOLUTION | | Descriptor: | 2-IMINOBIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-05 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NDJ
 
 | | Streptavidin Mutant Y43F with Biotin at 1.81A Resolution | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-09 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N43
 
 | | Streptavidin Mutant N23A with biotin at 1.89A | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QNZ
 
 | | NMR structure of the 0.5b anti-HIV antibody complex with the gp120 V3 peptide | | Descriptor: | 0.5B ANTIBODY (HEAVY CHAIN), 0.5B ANTIBODY (LIGHT CHAIN), GP120 | | Authors: | Tugarinov, V, Zvi, A, Levy, R, Hayek, Y, Matsushita, S, Anglister, J. | | Deposit date: | 1999-10-26 | | Release date: | 2000-06-03 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Anti-Gp120 Antibody Complex with a V3 Peptide Reveals a Surface Important for Co-Receptor Binding
Structure, 8, 2000
|
|
2DC5
 
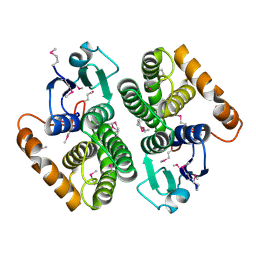 | | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution | | Descriptor: | Glutathione S-transferase, mu 7 | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-28 | | Release date: | 2006-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution
To be Published
|
|
2E3V
 
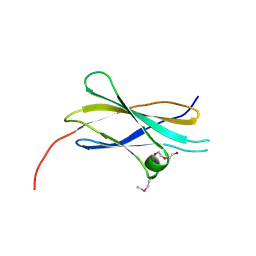 | | Crystal structure of the first fibronectin type III domain of neural cell adhesion molecule splicing isoform from human muscle culture lambda-4.4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nishino, A, Saijo, S, Kishishita, S, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-11-30 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the first fibronectin type III domain of neural cell adhesion molecule splicing isoform from human muscle culture lambda-4.4
To be Published
|
|
1RH8
 
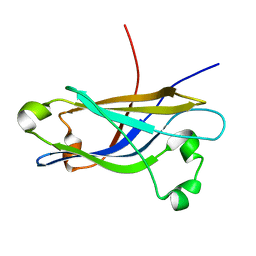 | | Three-dimensional structure of the calcium-free Piccolo C2A-domain | | Descriptor: | Piccolo protein | | Authors: | Garcia, J, Gerber, S.H, Sugita, S, Sudhof, T.C, Rizo, J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the Piccolo C2A domain regulated by alternative splicing.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2DYQ
 
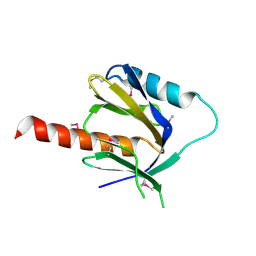 | | Crystal Structure of the C-terminal Phophotyrosine Interaction Domain of Human APBB3 | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 3 | | Authors: | Nishino, A, Saijo, S, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the C-terminal Phophotyrosine Interaction Domain of Human APBB3
To be Published
|
|
2D8N
 
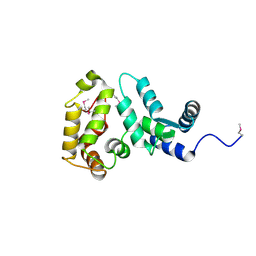 | | Crystal structure of human recoverin at 2.2 A resolution | | Descriptor: | Recoverin | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-07 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human recoverin at 2.2 A resolution
To be Published
|
|
1HR6
 
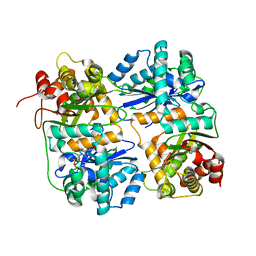 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
2D7U
 
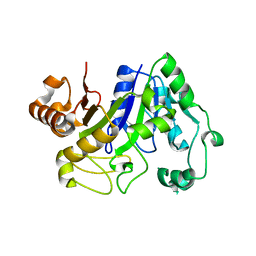 | | Crystal structure of hypothetical adenylosuccinate synthetase, PH0438 from Pyrococcus horikoshii OT3 | | Descriptor: | Adenylosuccinate synthetase | | Authors: | Xie, Y, Kishisita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-29 | | Release date: | 2006-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical adenylosuccinate synthetase, PH0438 from Pyrococcus horikoshii OT3
To be Published
|
|
1IVW
 
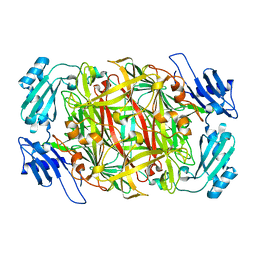 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVV
 
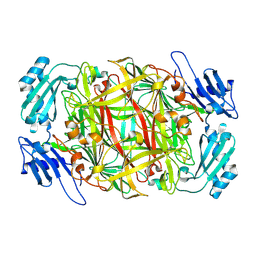 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVU
 
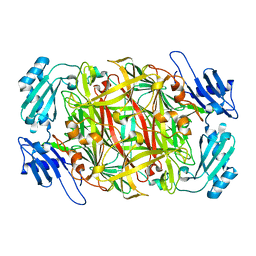 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
2E8A
 
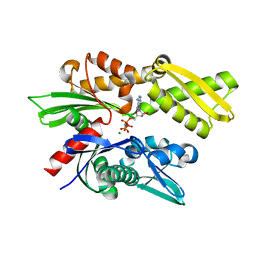 | | Crystal structure of the human Hsp70 ATPase domain in complex with AMP-PNP | | Descriptor: | Heat shock 70kDa protein 1A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Shida, M, Ishii, R, Takagi, T, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1IVX
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Holo form generated by biogenesis in crystal. | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1L1W
 
 | | NMR structure of a SRP19 binding domain in human SRP RNA | | Descriptor: | SRP19 binding domain of SRP RNA | | Authors: | Sakamoto, T, Morita, S, Tabata, K, Nakamura, K, Kawai, G. | | Deposit date: | 2002-02-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a SRP19 binding domain in human SRP RNA.
J.Biochem.(Tokyo), 132, 2002
|
|
