7TE5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis | | Descriptor: | MAGNESIUM ION, Pirin family protein Yhak | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis
To Be Published
|
|
7TG5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the Presence of Fe Ion from Yersinia pestis | | Descriptor: | CHLORIDE ION, FE (III) ION, Pirin family protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the presence of Fe ion from Yersinia pestis
To Be Published
|
|
7TFQ
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, FORMIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis
To Be Published
|
|
7THH
 
 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
7TEM
 
 | | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis
To Be Published
|
|
7TMU
 
 | | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function YPO0625 from Yersinia pestis
To Be Published
|
|
7TL5
 
 | | Crystal structure of putative hydrolase yjcS from Klebsiella pneumoniae. | | Descriptor: | 1,2-ETHANEDIOL, Lactamase_B domain-containing protein | | Authors: | Chang, C, Endres, M, Wu, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7TJ1
 
 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
7TWE
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DUF1479 domain-containing protein, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid
To Be Published
|
|
7TWC
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DUF1479 domain-containing protein, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS
To Be Published
|
|
6W01
 
 | | The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
6VYO
 
 | | Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6W61
 
 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
6W0P
 
 | | Putative kojibiose phosphorylase from human microbiome | | Descriptor: | Kojibiose phosphorylase | | Authors: | Dementiev, A, Osipiuk, J, Endres, M, Wakatsuki, S, Hess, M, Joachimiak, A. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Putative kojibiose phosphorylase from human microbiome
to be published
|
|
6U2Y
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, NICKEL (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions
To Be Published
|
|
6U0Z
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed penicillin G | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed penicillin G.
To Be Published
|
|
6U2Z
 
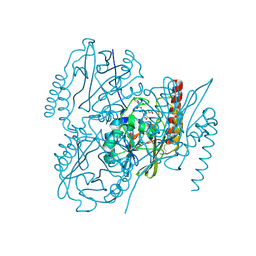 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, 1,2-ETHANEDIOL, COPPER (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions
To Be Published
|
|
6UAC
 
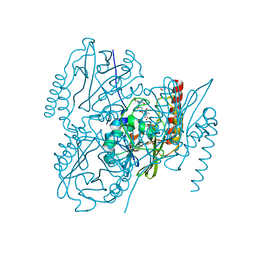 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with cadmium and hydrolyzed moxolactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, CADMIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with cadmium and hydrolyzed moxolactam
To Be Published
|
|
6U0I
 
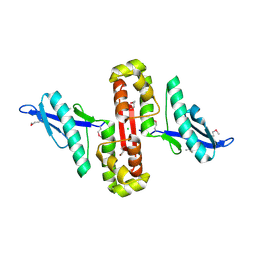 | |
6U0Y
 
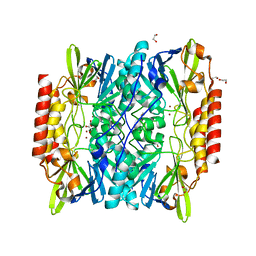 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia
To Be Published
|
|
6UA1
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase) | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form.
To Be Published
|
|
6UAF
 
 | | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem
To Be Published
|
|
6UR7
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | Descriptor: | Muramoyltetrapeptide carboxypeptidase | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
6V72
 
 | | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis
To Be Published
|
|
