5YNV
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, aromatic prenyltransferase | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
1G69
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 2-METHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-11-03 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
6K4Y
 
 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
7XFR
 
 | |
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
5YVK
 
 | | Crystal structure of a cyclase Famc1 from Fischerella ambigua UTEX 1903 | | Descriptor: | CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YVP
 
 | | Crystal structure of an apo form cyclase Filc1 from Fischerella sp. TAU | | Descriptor: | CALCIUM ION, TETRAETHYLENE GLYCOL, cyclase A | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z54
 
 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-17 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z53
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, SULFATE ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1G4P
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 4-AMINO-2-TRIFLUOROMETHYL-5-HYDROXYMETHYLPYRIMIDINE PYROPHOSPHATE, MAGNESIUM ION, THIAMIN PHOSPHATE SYNTHASE | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-27 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
5Z1W
 
 | |
1G6C
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-11-03 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
1G4E
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | THIAMIN PHOSPHATE SYNTHASE | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
1G4T
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 3-(4-AMINO-2-TRIFLUOROMETHYL-PYRIMIDIN-5-YLMETHYL)-4-METHYL-5-(2-PHOSPHONATOOXY-ETHYL)-THIAZOL-3-IUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-28 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
4J5I
 
 | |
6ITP
 
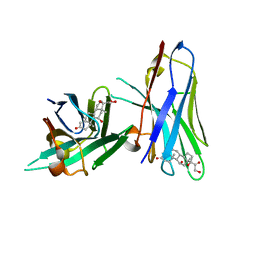 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
1G4S
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-27 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
1G67
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 2-METHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-11-03 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
1GN4
 
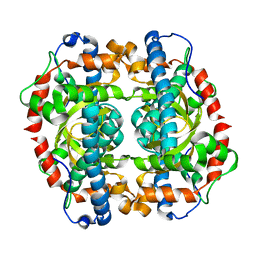 | | H145E mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants
Eur.J.Biochem., 251, 1998
|
|
1GN3
 
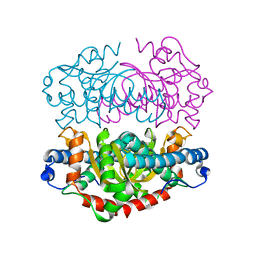 | | H145Q mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants.
Eur.J.Biochem., 251, 1998
|
|
1GN2
 
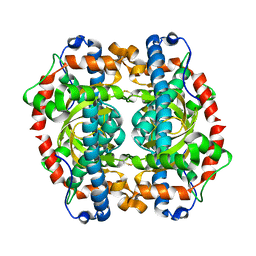 | | S123C mutant of the iron-superoxide dismutase from Mycobacterium tuberculosis. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Tickle, I.J, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Engineering of an Intersubunit Disulfide Bridge in the Iron-Superoxide Dismutase of Mycobacterium Tuberculosis.
Arch.Biochem.Biophys., 397, 2002
|
|
5YVL
 
 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7XSR
 
 | | Structure of Craspase-target RNA | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSQ
 
 | | Structure of the Craspase | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XUF
 
 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
