6KJ8
 
 | | E. coli ATCase holoenzyme mutant - G166P (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJA
 
 | | E. coli ATCase holoenzyme mutant - G128/130A (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.064 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
5J87
 
 | |
8ZJB
 
 | |
5GRB
 
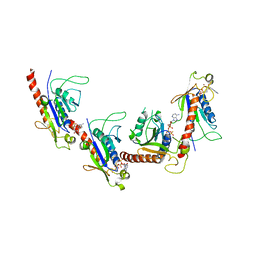 | | Crystal structure of 2C helicase from enterovirus 71 (EV71) bound with ATPgammaS | | Descriptor: | EV71 2C ATPase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Guan, H.X, Tian, J, Qin, B, Wojdyla, J, Wang, M.T, Cui, S. | | Deposit date: | 2016-08-09 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of 2C helicase from enterovirus 71
SCI ADV, 3, 2017
|
|
5GQ1
 
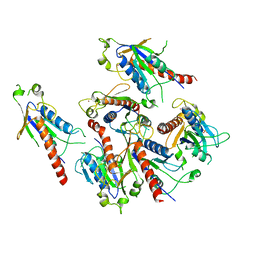 | | Crystal structure of 2C helicase from enterovirus 71 (EV71) | | Descriptor: | Genome polyprotein, PHOSPHATE ION, ZINC ION | | Authors: | Guan, H.X, Tian, J, Qin, B, Wojdyla, J, Wang, M.T, Cui, S. | | Deposit date: | 2016-08-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Crystal structure of 2C helicase from enterovirus 71
SCI ADV, 3, 2017
|
|
9IY1
 
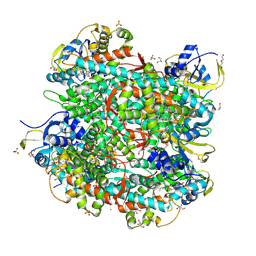 | | P450 BS beta mutant F46A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fatty-acid peroxygenase, GLYCEROL, ... | | Authors: | Gong, P.Q, Gao, X. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Unexpected Activities of CYP152 Peroxygenases towards Non-carboxylic Substrates Reveal Novel Substrate Recognition Mechanism and Catalytic Versatility.
Angew.Chem.Int.Ed.Engl., 2025
|
|
7M71
 
 | | SARS-CoV-2 Spike:5A6 Fab complex I focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 5A6 Fab heavy chain, Antibody 5A6 Fab light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
7M7B
 
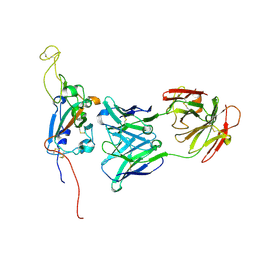 | | SARS-CoV-2 Spike:Fab 3D11 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 3D11 heavy chain, Antibody Fab 3D11 light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-316 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEO
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253H55L and COVOX-75 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEK
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
9MCZ
 
 | | Crystal structure of the transpeptidase domain of PBP2 from the Neisseria gonorrhoeae cephalosporin decreased susceptibility strain 35/02 in complex with boronate inhibitor VNRX-6884 | | Descriptor: | (3R)-3-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-phosphonophenyl)acetyl]amino}-2-hydroxy-3,4-dihydro-2H-1,2-benzoxaborinine-8-carboxylic acid, Penicillin-binding protein 2 | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A new class of penicillin-binding protein inhibitors to address drug-resistant Neisseria gonorrhoeae.
Biorxiv, 2024
|
|
9MD0
 
 | | Crystal structure of the transpeptidase domain of PBP2 from the Neisseria gonorrhoeae cephalosporin decreased susceptibility strain 35/02 in complex with boronate inhibitor VNRX-6752 | | Descriptor: | (3R)-3-({(2R)-2-(4-carboxyphenyl)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]acetyl}amino)-2-hydroxy-3,4-dihydro-2H-1,2-benzoxaborinine-8-carboxylic acid, Penicillin-binding protein 2 | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | A new class of penicillin-binding protein inhibitors to address drug-resistant Neisseria gonorrhoeae.
Biorxiv, 2024
|
|
7KQB
 
 | | SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 5A6 heavy chain, Fab 5A6 light chain, ... | | Authors: | Asarnow, D, Charles, C, Cheng, Y. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
7KQE
 
 | |
7ORB
 
 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7OR9
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7ORA
 
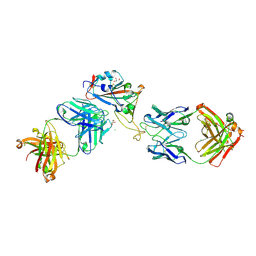 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7BP4
 
 | |
7BP6
 
 | |
7BP2
 
 | |
7BP5
 
 | |
