2QSY
 
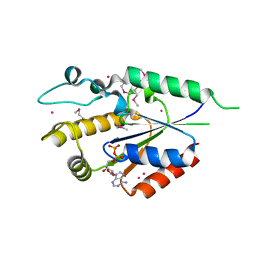 | | Human nicotinamide riboside kinase 1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2LXA
 
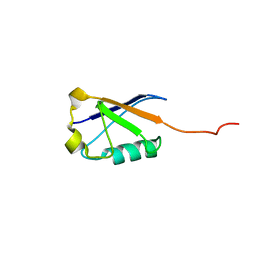 | |
2QNR
 
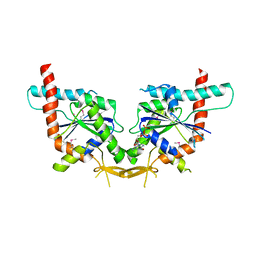 | | Human septin 2 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin-2, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Nedyalkova, L, Tempel, W, Landry, R, Crombet, L, Kozieradzki, I, Senisterra, G, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human septin 2 in complex with GDP.
To be Published
|
|
2QT1
 
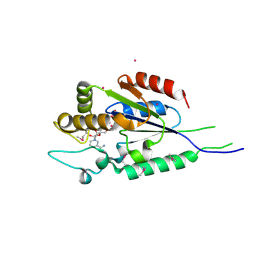 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide riboside | | Descriptor: | Nicotinamide riboside, Nicotinamide riboside kinase 1, PHOSPHATE ION, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2LXB
 
 | |
2LXC
 
 | |
2NZT
 
 | | Crystal structure of human hexokinase II | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Hexokinase-2, UNKNOWN ATOM OR ION, ... | | Authors: | Rabeh, W.M, Zhu, H, Nedyalkova, L, Tempel, W, Wasney, G, Landry, R, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-25 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation.
Biosci.Rep., 38, 2018
|
|
2PF0
 
 | |
2PZA
 
 | | NAD+ Synthetase from Bacillus anthracis with AMP + PPi and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | McDonald, H.M, Pruett, P.S, Deivanayagam, C, Protasevich, I.I, Carson, W.M, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD(+) synthetase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2Q3F
 
 | | X-ray crystal structure of putative human Ras-related GTP binding D in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related GTP-binding protein D | | Authors: | Mulichak, A.M, Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Keefe, L.J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Ras-related GTP-binding D.
To be published
|
|
2LO0
 
 | |
2PZ8
 
 | | NAD+ Synthetase from Bacillus anthracis with AMP-CPP and Mg2+ | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | McDonald, H.M, Pruett, P.S, Deivanayagam, C, Protasevich, I.I, Carson, W.M, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD(+) synthetase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QSZ
 
 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide mononucleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, CHLORIDE ION, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2QT0
 
 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide riboside and an ATP analogue | | Descriptor: | MAGNESIUM ION, Nicotinamide riboside, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2TMG
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT S128R, T158E, N117R, S160E | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, van der Oost, J, de Vos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. II: construction of a 16-residue ion-pair network at the subunit interface.
J.Mol.Biol., 289, 1999
|
|
2PBO
 
 | |
1RGQ
 
 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | Descriptor: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | Authors: | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
1ROC
 
 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
1QB2
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED SUBDOMAIN OF HUMAN PROTEIN SRP54M AT 2.1A RESOLUTION: EVIDENCE FOR THE MECHANISM OF SIGNAL PEPTIDE BINDING | | Descriptor: | HUMAN SIGNAL RECOGNITION PARTICLE 54 KD PROTEIN | | Authors: | Clemons Jr, W.M, Gowda, K, Black, S.D, Zwieb, C, Ramakrishnan, V. | | Deposit date: | 1999-04-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 A resolution: evidence for the mechanism of signal peptide binding.
J.Mol.Biol., 292, 1999
|
|
1RH5
 
 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
1R6V
 
 | | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin | | Descriptor: | CALCIUM ION, subtilisin-like serine protease | | Authors: | Kim, J.S, Kluskens, L.D, de Vos, W.M, Huber, R, van der Oost, J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin.
J.Mol.Biol., 335, 2004
|
|
1TF2
 
 | |
1T1L
 
 | | Crystal structure of the long-chain fatty acid transporter FadL | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1TF5
 
 | |
1TM6
 
 | |
