3W7S
 
 | | Escherichia coli K12 YgjK complexed with glucose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-glucopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3W7U
 
 | | Escherichia coli K12 YgjK complexed with galactose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-galactopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3G5V
 
 | | Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR | | Descriptor: | 806 light chain, 808 heavy chain, ACETATE ION, ... | | Authors: | Garrett, T.P.J, Burgess, A.W, Huyton, T, Xu, Y. | | Deposit date: | 2009-02-05 | | Release date: | 2010-02-09 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Antibodies specifically targeting a locally misfolded region of tumor associated EGFR
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KK8
 
 | | NMR Solution Structure of a Putative Uncharacterized Protein obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium target AR3449A | | Descriptor: | Uncharacterized protein AT4g05270 | | Authors: | Mani, R, Gurla, S.V.T, Shastry, R, Ciccosanti, C, Foote, E, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y, Acton, T, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Putative Uncharacterized Protein obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium Target AR3449A
To be Published
|
|
2KJ6
 
 | | NMR Solution Structure of a Tubulin folding cofactor B obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium target AR3436A | | Descriptor: | Tubulin folding cofactor B | | Authors: | Mani, R, Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y.J, Acton, T, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Tbulin folding Cofactor B obtained from Arabidopsis thaliana: Northeast
To be Published
|
|
2KPM
 
 | | Solution NMR Structure of uncharacterized protein from gene locus NE0665 of Nitrosomonas europaea. Northeast Structural Genomics Target NeR103A | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Belote, R, Jiang, M, Xiao, R, Ciccosanti, C, Acton, T, Everett, J, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of uncharacterized protein from gene locus NE0665 of Nitrosomonas europaea. Northeast Structural Genomics Target NeR103A
To be Published
|
|
2KVT
 
 | | solution NMR structure of yaiA from Escherichia Eoli. Northeast Structural Genomics Target ER244 | | Descriptor: | Uncharacterized protein yaiA | | Authors: | Tang, Y, Chen, X, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-28 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | solution NMR structure of yaiA from Escherichia coli. Northeast Structural Genomics Target ER244
To be Published
|
|
2KRT
 
 | | Solution NMR Structure of a Conserved Hypothetical Membrane Lipoprotein Obtained from Ureaplasma parvum: Northeast Structural Genomics Consortium Target UuR17A (139-239) | | Descriptor: | Conserved hypothetical membrane lipoprotein | | Authors: | Mani, R, Swapna, G, Janjua, H, Ciccosanti, C, Huang, Y, Patel, D, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Conserved Hypothetical Membrane Lipoprotein obtained from Ureaplasma parvum : Northeast Structural Genomics Consortium Target UuR17A (139-239)
To be Published
|
|
2F6K
 
 | | Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LpR24 | | Descriptor: | MANGANESE (II) ION, metal-dependent hydrolase | | Authors: | Das, K, Xiao, R, Acton, T, Ma, L, Arnold, E, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-29 | | Release date: | 2006-01-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of ACMDS from Lactobacillus plantarum
To be Published
|
|
2LFC
 
 | | Solution NMR Structure of Fumarate reductase flavoprotein subunit from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR145J | | Descriptor: | Fumarate reductase, flavoprotein subunit | | Authors: | Liu, G, Tang, Y, Xiao, R, Janjua, H, Ciccosanti, C, Wang, H, Acton, T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target LpR145J
To be Published
|
|
2D0F
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D0H
 
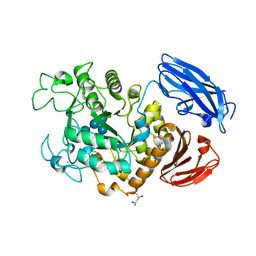 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2BN8
 
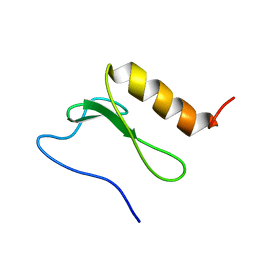 | | Solution Structure and interactions of the E .coli Cell Division Activator Protein CedA | | Descriptor: | CELL DIVISION ACTIVATOR CEDA | | Authors: | Chen, H.A, Simpson, P, Huyton, T, Roper, D, Matthews, S. | | Deposit date: | 2005-03-22 | | Release date: | 2006-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Biochemistry, 44, 2005
|
|
2D2O
 
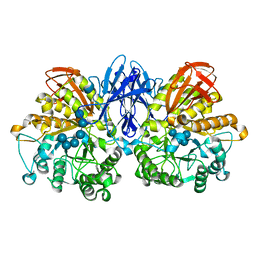 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
2D0G
 
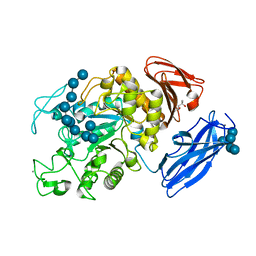 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2EW0
 
 | | X-ray Crystal Structure of Protein Q6FF54 from Acinetobacter sp. ADP1. Northeast Structural Genomics Consortium Target AsR1. | | Descriptor: | SULFATE ION, hypothetical protein ACIAD0353 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Forouhar, F, Acton, T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel x-ray structure of hypothetical protein Q6FF54 at the resolution 1.4 A. Northeast Structural Genomics target AsR1.
TO BE PUBLISHED
|
|
1IZJ
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1 mutant enzyme f313a | | Descriptor: | CALCIUM ION, amylase | | Authors: | Ohtaki, A, Iguchi, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual conversion of substrate specificities of Thermoactinomyces vulgaris R-47 alpha-amylases TVAI and TVAII by site-directed mutagenesis
CARBOHYDR.RES., 338, 2003
|
|
1IZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1 mutant enzyme w398v | | Descriptor: | CALCIUM ION, amylase | | Authors: | Ohtaki, A, Iguchi, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual conversion of substrate specificities of Thermoactinomyces vulgaris R-47 alpha-amylases TVAI and TVAII by site-directed mutagenesis
CARBOHYDR.RES., 338, 2003
|
|
1IY9
 
 | | Crystal structure of spermidine synthase | | Descriptor: | Spermidine synthase | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-07-26 | | Release date: | 2002-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of spermidine synthase
To be Published
|
|
1JI1
 
 | | Crystal Structure Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 | | Descriptor: | ALPHA-AMYLASE I, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
1LUR
 
 | | Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66 | | Descriptor: | SULFATE ION, aldose 1-epimerase | | Authors: | Keller, J.P, Xiao, R, MacDonald, L, Shen, J, Acton, T, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-23 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the GalM/aldose Epimerase Homologue from C. elegans, Northeast Structural Genomics Target WR66
TO BE PUBLISHED
|
|
1M0S
 
 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID IR21) | | Descriptor: | CITRIC ACID, Ribose-5-Phosphate Isomerase A | | Authors: | Das, K, Xiao, R, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-14 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-RIBOSE-5-PHOSPHATE ISOMERASE, IR21
TO BE PUBLISHED
|
|
1WZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WMR
 
 | | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
