4NKQ
 
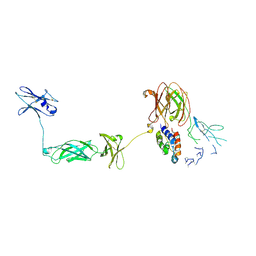 | | Structure of a Cytokine Receptor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Parker, M.W, Broughton, S.E. | | Deposit date: | 2013-11-13 | | Release date: | 2015-09-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Conformational Changes in the GM-CSF Receptor Suggest a Molecular Mechanism for Affinity Conversion and Receptor Signaling.
Structure, 24, 2016
|
|
6HCV
 
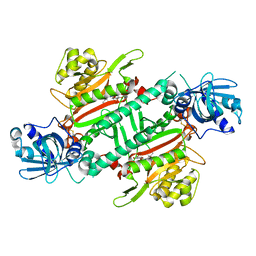 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with a chromone ligand | | Descriptor: | 6-fluoranyl-~{N}-[(1-oxidanylcyclohexyl)methyl]-4-oxidanylidene-chromene-2-carboxamide, Lysine--tRNA ligase | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1OM1
 
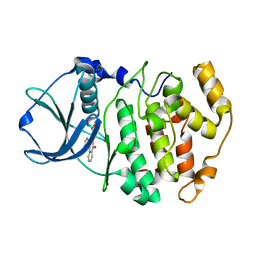 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
2JW6
 
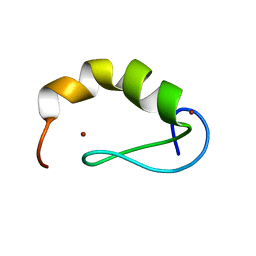 | | Solution structure of the DEAF1 MYND domain | | Descriptor: | Deformed epidermal autoregulatory factor 1 homolog, ZINC ION | | Authors: | Spadaccini, R, Perrin, H, Bottomley, M, Ansieu, S, Sattler, M. | | Deposit date: | 2007-10-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508].
J.Mol.Biol., 376, 2008
|
|
6FBV
 
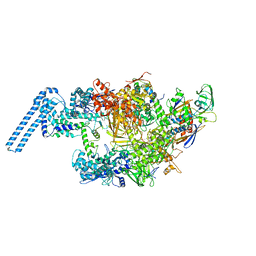 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Das, K, Lin, W, Ebright, E. | | Deposit date: | 2017-12-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
4APN
 
 | | Structure of TR from Leishmania infantum in complex with a diarylpirrole-based inhibitor | | Descriptor: | 4-[[1-(4-ethylphenyl)-2-methyl-5-(4-methylsulfanylphenyl)pyrrol-3-yl]methyl]thiomorpholine, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G, Biava, M. | | Deposit date: | 2012-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Leishmania Infantum Trypanothione Reductase by Azole-Based Compounds: A Comparative Analysis with its Physiological Substrate by X-Ray Crystallography.
Chemmedchem, 8, 2013
|
|
6XQA
 
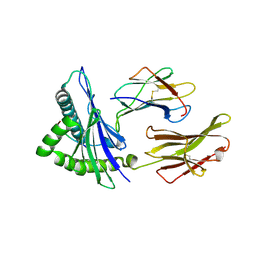 | | Crystal Structure of HLA A*2402 in complex with TYQWVLKNL, an 9-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
1BYX
 
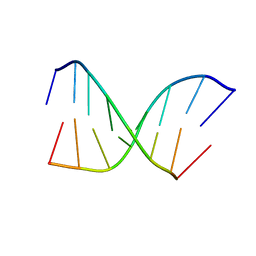 | | CHIMERIC HYBRID DUPLEX R(GCAGUGGC).R(GCCA)D(CTGC) COMPRISING THE TRNA-DNA JUNCTION FORMED DURING INITIATION OF HIV-1 REVERSE TRANSCRIPTION | | Descriptor: | DNA/RNA (5'-R(*GP*CP*CP*A)-D(P*CP*TP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*UP*GP*GP*C)-3') | | Authors: | Szyperski, T, Goette, M, Billeter, M, Perola, E, Cellai, L. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription.
J.Biomol.NMR, 13, 1999
|
|
8R3E
 
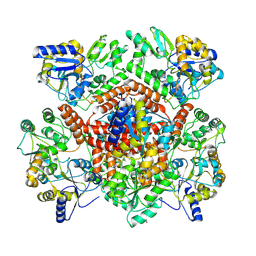 | | Huntingtin, 1-17, MBP-N | | Descriptor: | Maltodextrin-binding protein, ZINC ION | | Authors: | Steinbacher, S, Toledo-Sherman, L, Dominguez, C. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Post-translational modifications in the first 17 amino acids of huntingtin influence self-association and interaction with membranes
To Be Published
|
|
8R2O
 
 | | Huntingtin-Q17, 1-66, N-MBP fusion | | Descriptor: | Maltodextrin-binding protein,Huntingtin, myristoylated N-terminal fragment, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Toledo-Sherman, L, Dominguez, C. | | Deposit date: | 2023-11-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Post-translational modifications in the first 17 amino acids of huntingtin influence self-association and interaction with membranes
To Be Published
|
|
8QFT
 
 | |
5GVP
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS654 | | Descriptor: | 3-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]phenyl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
6G8G
 
 | | Flavonoid-responsive Regulator FrrA in complex with Genistein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GENISTEIN, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
6NMG
 
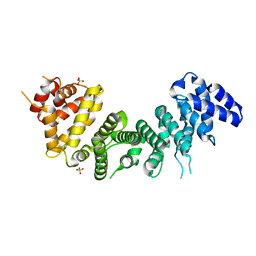 | | Crystal Structure of Rat Ric-8A G alpha binding domain | | Descriptor: | Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans), SULFATE ION | | Authors: | Zeng, B, Mou, T.C, Sprang, S.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Structure, 27, 2019
|
|
5GVN
 
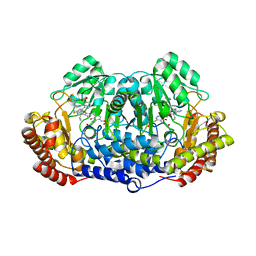 | | Plasmodium vivax SHMT bound with PLP-glycine and GS653 | | Descriptor: | 3-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]phenyl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
5GVL
 
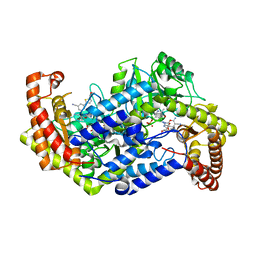 | | Plasmodium vivax SHMT bound with PLP-glycine and GS182 | | Descriptor: | (4~{S})-6-azanyl-4-[3-cyano-5-[5-(methoxymethyl)thiophen-2-yl]phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
5GVM
 
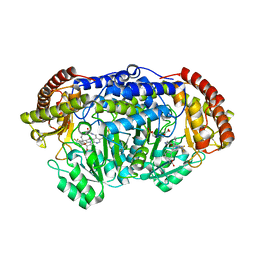 | | Plasmodium vivax SHMT bound with PLP-glycine and GS557 | | Descriptor: | 2-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]phenyl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
5GVK
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS256 | | Descriptor: | 5-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-cyano-phenyl]-~{N},~{N}-dimethyl-thiophene-2-sulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
6G8H
 
 | | Flavonoid-responsive Regulator FrrA in complex with (R,S)-Naringenin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, NARINGENIN, R-naringenin, ... | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
6NMJ
 
 | | Crystal Structure of Rat Ric-8A G alpha binding domain, "Paratone-N Immersed" | | Descriptor: | Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Zeng, B, Mou, T.C, Sprang, S.R. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Structure, 27, 2019
|
|
6NT2
 
 | | type 1 PRMT in complex with the inhibitor GSK3368715 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, N~1~-({5-[4,4-bis(ethoxymethyl)cyclohexyl]-1H-pyrazol-4-yl}methyl)-N~1~,N~2~-dimethylethane-1,2-diamine, ... | | Authors: | Concha, N.O. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss.
Cancer Cell, 36, 2019
|
|
4ADW
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA INFANTUM TRYPANOTHIONE REDUCTASE IN COMPLEX WITH NADPH AND TRYPANOTHIONE | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G, Malatesta, F, Fiorillo, A. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Inhibition of Leishmania Infantum Trypanothione Reductase by Azole-Based Compounds: A Comparative Analysis with its Physiological Substrate by X-Ray Crystallography.
Chemmedchem, 8, 2013
|
|
8SXU
 
 | |
8SXT
 
 | | Structure of LINE-1 ORF2p with template:primer hybrid | | Descriptor: | DNA primer, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | van Eeuwen, T, Taylor, M.S, Rout, M.P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
7SR6
 
 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
