7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
1UDY
 
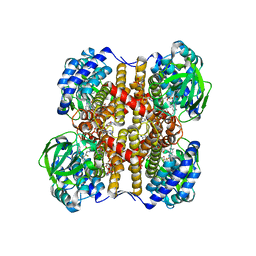 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
2YYV
 
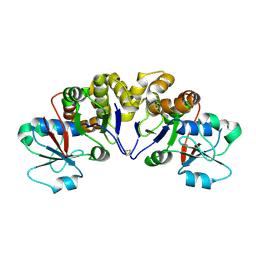 | | Crystal structure of uncharacterized conserved protein from Thermotoga maritima | | Descriptor: | Probable 2-phosphosulfolactate phosphatase | | Authors: | Nakagawa, N, Nakamura, Y, Bessho, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermotoga maritima
To be Published
|
|
2N37
 
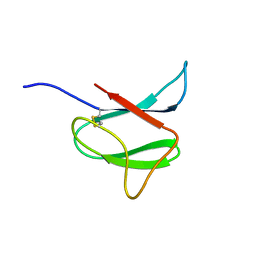 | | Solution structure of AVR-Pia | | Descriptor: | AVR-Pia protein | | Authors: | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
2E0Z
 
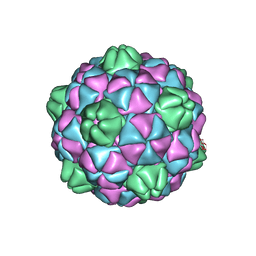 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
3GP7
 
 | |
5GHI
 
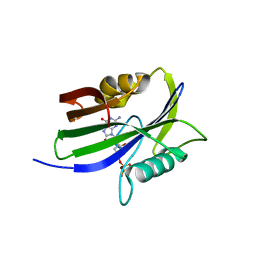 | | Crystal structure of human MTH1(G2K mutant) in complex with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHO
 
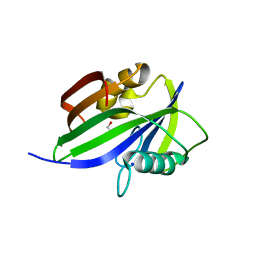 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHN
 
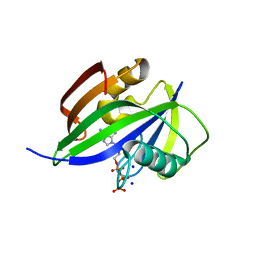 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHM
 
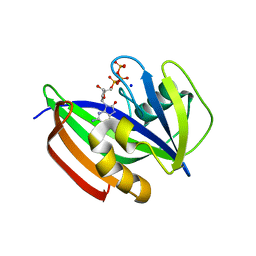 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 7.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHQ
 
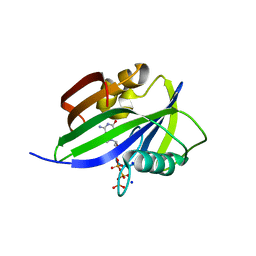 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 2-oxo-dATP under high concentrations of 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.181 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHJ
 
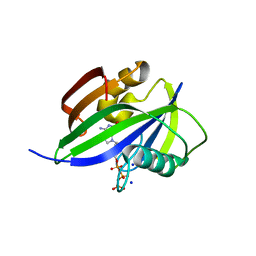 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHP
 
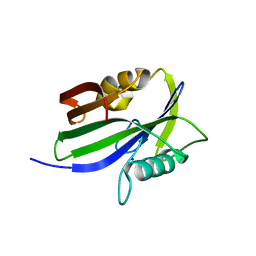 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
2EK2
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (E140M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Tanaka, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (E140M)
To be Published
|
|
2PA0
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sugahara, M, Tanaka, Y, Matsuura, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2P75
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL, SODIUM ION | | Authors: | Sugahara, M, Tanaka, Y, Matsuura, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2OWG
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Sugahara, M, Matsuura, Y, Tanaka, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-16 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
1ERZ
 
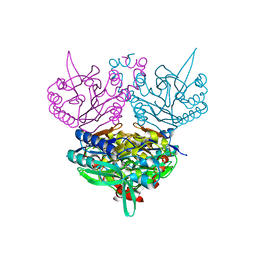 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | Descriptor: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | Authors: | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | Deposit date: | 2000-04-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ5
 
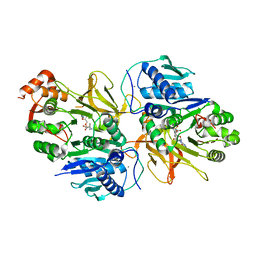 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
3VZR
 
 | |
5IX7
 
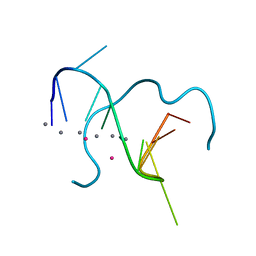 | | Crystal structure of metallo-DNA nanowire with infinite one-dimensional silver array | | Descriptor: | DNA (5'-D(*GP*GP*AP*CP*TP*(CBR)P*GP*AP*CP*TP*CP*C)-3'), POTASSIUM ION, SILVER ION | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Hattori, Y, Saneyoshi, H, Ono, A, Tanaka, Y. | | Deposit date: | 2016-03-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A metallo-DNA nanowire with uninterrupted one-dimensional silver array
Nat Chem, 9, 2017
|
|
6AIT
 
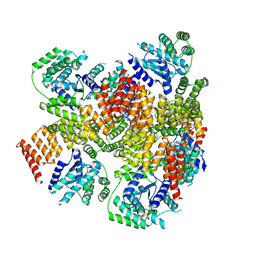 | | Crystal structure of E. coli BepA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-barrel assembly-enhancing protease, ZINC ION | | Authors: | Umar, M.S.M, Tanaka, Y, Kamikubo, H, Tsukazaki, T. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Function of the beta-Barrel Assembly-Enhancing Protease BepA.
J. Mol. Biol., 431, 2019
|
|
