5ZAS
 
 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5ZBZ
 
 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
6CFD
 
 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
3JVR
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
3JVS
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | 2-[(4-tert-butyl-3-nitrophenyl)carbonyl]-N-naphthalen-1-ylhydrazinecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
5GON
 
 | | Structures of a beta-lactam bridged analogue in complex with tubulin | | Descriptor: | (3R,4R)-4-(4-methoxy-3-oxidanyl-phenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhou, L, Liu, Y, Cheng, L, Wang, Y. | | Deposit date: | 2016-07-28 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Potent Antitumor Activities and Structure Basis of the Chiral beta-Lactam Bridged Analogue of Combretastatin A-4 Binding to Tubulin.
J. Med. Chem., 59, 2016
|
|
8WOY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOZ
 
 | | Cryo-EM structure of SARS-CoV RBD in complex with rabbit ACE2 | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOX
 
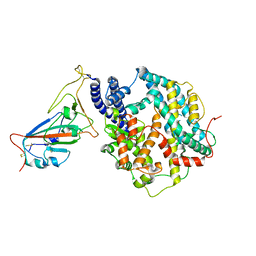 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8XZB
 
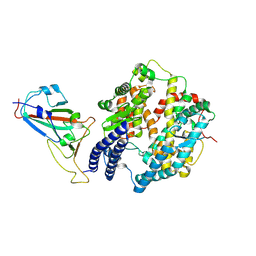 | | The structure of fox ACE2 and SARS-CoV RBD complex | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
8XZD
 
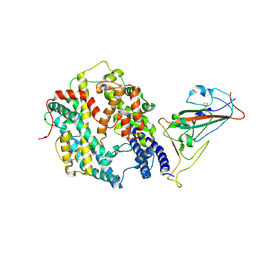 | | The structure of fox ACE2 and Omicron BF.7 RBD complex | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-21 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
8XYZ
 
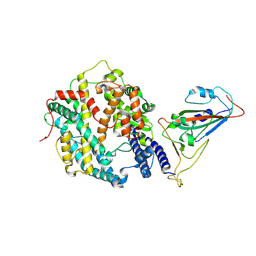 | | The structure of fox ACE2 and PT RBD complex | | Descriptor: | Angiotensin-converting enzyme, Signal peptide, Spike protein S1, ... | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
8KER
 
 | |
8KDT
 
 | |
8KDS
 
 | |
8KEP
 
 | |
8KDM
 
 | |
8KDR
 
 | |
8KEO
 
 | |
8KEK
 
 | |
8KEQ
 
 | |
8KEJ
 
 | |
8KEH
 
 | |
3DM7
 
 | |
8G1T
 
 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P21 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-02-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
