4X1N
 
 | | The crystal structure of mupain-1-16 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
5I4V
 
 | | Discovery of novel, orally efficacious Liver X Receptor (LXR) beta agonists | | Descriptor: | Oxysterols receptor LXR-beta,Nuclear receptor coactivator 2, Retinoic acid receptor RXR-beta,Nuclear receptor coactivator 2, {2-[(2R)-4-[4-(hydroxymethyl)-3-(methylsulfonyl)phenyl]-2-(propan-2-yl)piperazin-1-yl]-4-(trifluoromethyl)pyrimidin-5-yl}methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of a Novel, Orally Efficacious Liver X Receptor (LXR) beta Agonist.
J.Med.Chem., 59, 2016
|
|
8XVJ
 
 | | Cryo-EM structure of ETAR bound with Macitentan | | Descriptor: | 6-[2-(5-bromanylpyrimidin-2-yl)oxyethoxy]-5-(4-bromophenyl)-~{N}-(propylsulfamoyl)pyrimidin-4-amine, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVK
 
 | | Cryo-EM structure of ETAR bound with Ambrisentan | | Descriptor: | (2~{S})-2-(4,6-dimethylpyrimidin-2-yl)oxy-3-methoxy-3,3-diphenyl-propanoic acid, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVL
 
 | | Cryo-EM structure of ETAR bound with Zibotentan | | Descriptor: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVI
 
 | | Cryo-EM structure of ETAR bound with Endothelin1 | | Descriptor: | Endoglucanase H,Endothelin-1 receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVH
 
 | | Cryo-EM structure of ETBR bound with Endothelin1 | | Descriptor: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVE
 
 | | Cryo-EM structure of ETBR bound with BQ3020 | | Descriptor: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
4WI8
 
 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation Y436A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, ... | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
4WI2
 
 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc (wild-type) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
4WI7
 
 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation H435A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
4WI5
 
 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation H310A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
8X1N
 
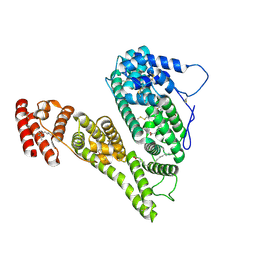 | | Cryo-EM structure of human alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein, PALMITIC ACID, ZINC ION, ... | | Authors: | Liu, Z.M, Li, M.S, Wu, C, Liu, K. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural characteristics of alpha-fetoprotein, including N-glycosylation, metal ion and fatty acid binding sites.
Commun Biol, 7, 2024
|
|
8XC4
 
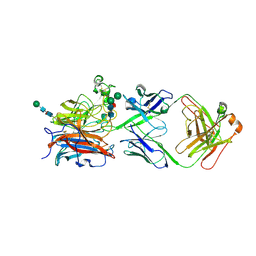 | | Nipah virus attachment glycoprotein head domain in complex with a broadly neutralizing antibody 1E5 | | Descriptor: | 1E5-VH, 1E5-VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, P.F, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A potent Henipavirus cross-neutralizing antibody reveals a dynamic fusion-triggering pattern of the G-tetramer.
Nat Commun, 15, 2024
|
|
8WI2
 
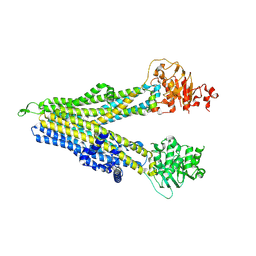 | | RD-hMRP5-inward open | | Descriptor: | ATP-binding cassette sub-family C member 5, CYS-GLN-ASP-ALA-LEU-GLU-THR-ALA-ALA-ARG-ALA-GLU-GLY-LEU-SER-LEU-ASP-ALA-SER | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI3
 
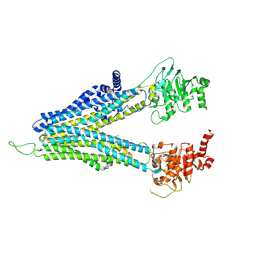 | | ND-hMRP5-inward open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8WI4
 
 | | m6-hMRP5 inward open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
9C4M
 
 | | Crystal Structure of A. baumannii GuaB dCBS with inhibitor G6 (3826) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(morpholin-4-yl)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Differential effects of inosine monophosphate dehydrogenase (IMPDH/GuaB) inhibition in Acinetobacter baumannii and Escherichia coli.
J.Bacteriol., 2024
|
|
8ZU2
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8g | | Descriptor: | 2-azanyl-5-[2-(1,4-diazepan-1-yl)pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, GLYCINE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.79888582 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8WI0
 
 | | wt-hMRP5 inward-open | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
8ZTX
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 6b | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-pyridin-4-yl-benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.70033228 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZUD
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8f | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-(2-morpholin-4-ylpyridin-4-yl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.50510085 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZUL
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8m | | Descriptor: | 2-azanyl-5-[2-[(3~{R})-3-azanylpyrrolidin-1-yl]pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.80026162 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8WI5
 
 | | M5PI-bound hMRP5 | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
7YTO
 
 | |
