159D
 
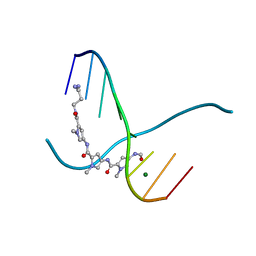 | | SIDE BY SIDE BINDING OF TWO DISTAMYCIN A DRUGS IN THE MINOR GROOVE OF AN ALTERNATING B-DNA DUPLEX | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*IP*CP*IP*CP*IP*CP*IP*C)-3'), MAGNESIUM ION | | Authors: | Chen, X, Ramakrishnan, B, Rao, S.T, Sundaralingam, M. | | Deposit date: | 1994-02-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Nat.Struct.Biol., 1, 1994
|
|
2VEK
 
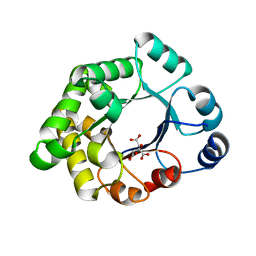 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | 3-(BUTYLSULPHONYL)-PROPANOIC ACID, CITRIC ACID, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEN
 
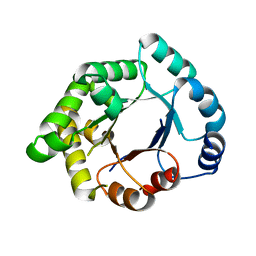 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | CITRIC ACID, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEI
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
217D
 
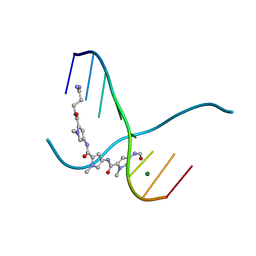 | |
216D
 
 | |
2VEM
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | (3-bromo-2-oxo-propoxy)phosphonic acid, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
2VEL
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties. | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
195D
 
 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
213D
 
 | |
1DQH
 
 | |
260D
 
 | |
246D
 
 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
251D
 
 | |
279D
 
 | |
1DQF
 
 | |
1DVL
 
 | |
2AWE
 
 | |
1VTF
 
 | |
7VRT
 
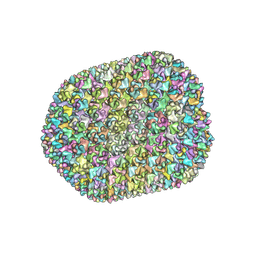 | | The unexpanded head structure of phage T4 | | Descriptor: | Capsid vertex protein, Major capsid protein | | Authors: | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VS5
 
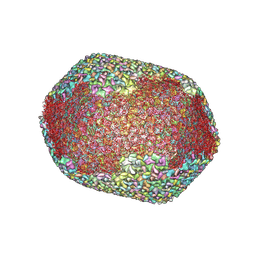 | | The expanded head structure of phage T4 | | Descriptor: | Capsid vertex protein, Major capsid protein, Small outer capsid protein | | Authors: | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2021-10-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1SPQ
 
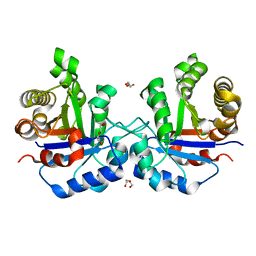 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SQ7
 
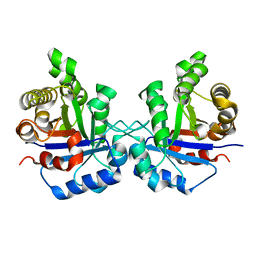 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1R41
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R3Z
 
 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*(5CM)P*GP*(5CM)P*GP*CP*GP*C)-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
