1TIY
 
 | | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160 | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Kuzin, A.P, Vorobiev, S, Edstrom, W, Forouhar, F, Acton, T, Shastry, R, Ma, L.-C, Chiang, Y.-W, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS
NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160
To be published
|
|
1T17
 
 | | Solution Structure of the 18 kDa Protein CC1736 from Caulobacter crescentus: The Northeast Structural Genomics Consortium Target CcR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Shen, Y, Atreya, H.S, Acton, T, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity.
Proteins, 58, 2005
|
|
2LFC
 
 | | Solution NMR Structure of Fumarate reductase flavoprotein subunit from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR145J | | Descriptor: | Fumarate reductase, flavoprotein subunit | | Authors: | Liu, G, Tang, Y, Xiao, R, Janjua, H, Ciccosanti, C, Wang, H, Acton, T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target LpR145J
To be Published
|
|
2ZYO
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with maltotetraose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
2ZYM
 
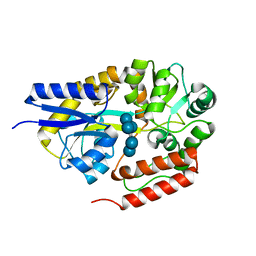 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
2ZYN
 
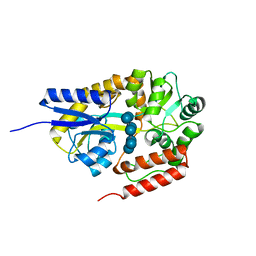 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with beta-cyclodextrin | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Matsumoto, M, Yamada, M, Kurakata, Y, Yoshida, H, Kamitori, S, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of open and closed forms of cyclo/maltodextrin-binding protein
Febs J., 276, 2009
|
|
3A6O
 
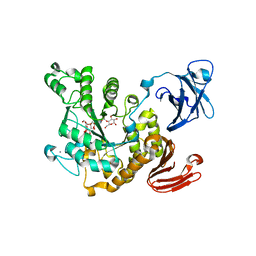 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/acarbose complex | | Descriptor: | ACARBOSE DERIVED PENTASACCHARIDE, CALCIUM ION, Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2009-09-07 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1XHJ
 
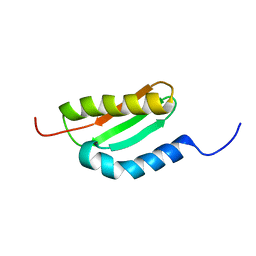 | | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630. Northest Structural Genomics Consortium Target SeR8. | | Descriptor: | Nitrogen Fixation Protein NifU | | Authors: | Baran, M.C, Huang, Y.P, Acton, T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of The Staphylococcus Epidermidis Protein SE0630.
Northest Strucutral Genomics Consortium Target SeR8.
To be Published
|
|
1XQB
 
 | | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC)target IR47. | | Descriptor: | Hypothetical UPF0066 protein HI0510 | | Authors: | Benach, J, Lee, I, Forouhar, F, Kuzin, A.P, Keller, J.P, Itkin, A, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC) target IR47.
To be Published
|
|
1JIB
 
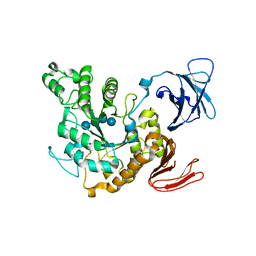 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
2L8O
 
 | | Solution structure of Chr148 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target Chr148 | | Descriptor: | Activator of Hsp90 ATPase homologue 1-like C-terminal domain-containing protein | | Authors: | Liu, Y, Lee, D, Ciccosanti, C, Nair, L, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Chr148 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target Chr148
To be Published
|
|
2LX7
 
 | | Solution NMR structure of SH3 domain of growth arrest-specific protein 7 (GAS7) (fragment 1-60) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8574A | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Yang, Y, Ramelot, T.A, Dan, L, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain of growth arrest-specific protein 7 (GAS7) (fragment 1-60) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8574A
To be Published
|
|
2LV2
 
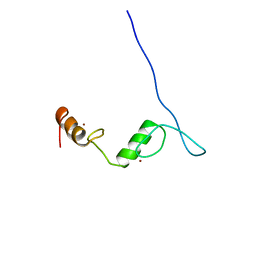 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
1JF6
 
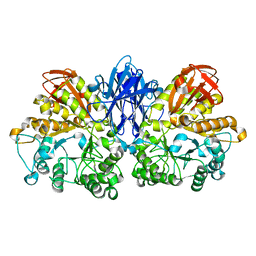 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JF5
 
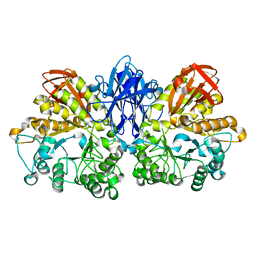 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JI2
 
 | | Improved X-ray Structure of Thermoactinomyces vulgaris R-47 alpha-Amylase 2 | | Descriptor: | ALPHA-AMYLASE II, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
2B95
 
 | | Solution NMR structure of protein dynein light chain 2A, cytoplasmic; Northeast structural genomics consortium TARGET HR2106 | | Descriptor: | Dynein light chain 2A | | Authors: | Liu, G, Atreya, H.S, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein dynein light chain 2A, cytoplasmic; Northeast structural genomics consortium TARGET HR2106
To be Published
|
|
2B6E
 
 | | X-Ray Crystal Structure of Protein HI1161 from Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR63. | | Descriptor: | ACETIC ACID, Hypothetical UPF0152 protein HI1161 | | Authors: | Kuzin, A.P, Benach, J, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Cunningham, K.E, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-01 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the different crystal form of the hypothetical UPF0152 protein HI1161: NESG target IR63
TO BE PUBLISHED
|
|
2AJA
 
 | | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR21. | | Descriptor: | ankyrin repeat family protein | | Authors: | Kuzin, A.P, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, C, Kellie, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila.
To be Published
|
|
2AJ2
 
 | | X-Ray Crystal Structure of Protein VC0467 from Vibrio cholerae. Northeast Structural Genomics Consortium Target VcR8. | | Descriptor: | Hypothetical UPF0301 protein VC0467 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | X-Ray structure of hypothetical protein VC0467 from Vibrio cholerae: new fold. Northeast Structural Genomics Consortium target VcR8.
To be Published
|
|
1UG9
 
 | | Crystal Structure of Glucodextranase from Arthrobacter globiformis I42 | | Descriptor: | CALCIUM ION, GLYCEROL, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Ohtaki, A, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1JL8
 
 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1UH3
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase/acarbose complex | | Descriptor: | (1S,2S,3R,6R)-6-amino-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-23 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
1UH2
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase/malto-hexaose complex | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-23 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
1UH4
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1/malto-tridecaose complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
