2JNV
 
 | | Solution structure of C-terminal domain of NifU-like protein from Oryza sativa | | Descriptor: | NifU-like protein 1, chloroplast | | Authors: | Saio, T, Ogura, K, Kumeta, H, Yokochi, M, Katoh, S, Katoh, E, Inagaki, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The cooperative role of OsCnfU-1A domain I and domain II in the iron sulphur cluster transfer process as revealed by NMR
J.Biochem.(Tokyo), 142, 2007
|
|
1IQ8
 
 | | Crystal Structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii | | Descriptor: | ARCHAEOSINE TRNA-GUANINE TRANSGLYCOSYLASE, MAGNESIUM ION, ZINC ION | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IO8
 
 | | Thermophilic cytochrome P450 (CYP119) from sulfolobus solfataricus: High resolution structural origin of its thermostability and functional properties | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties
J.Inorg.Biochem., 91, 2002
|
|
1J09
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP and Glu | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTAMIC ACID, Glutamyl-tRNA synthetase, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
1IO9
 
 | | THERMOPHILIC CYTOCHROME P450 (CYP119) FROM SULFOLOBUS SOLFATARICUS: HIGH RESOLUTION STRUCTURAL ORIGIN OF ITS THERMOSTABILITY AND FUNCTIONAL PROPERTIES | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties.
J.Inorg.Biochem., 91, 2002
|
|
1IOZ
 
 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1IQ0
 
 | | THERMUS THERMOPHILUS ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGINYL-TRNA SYNTHETASE | | Authors: | Shimada, A, Nureki, O, Goto, M, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J1U
 
 | | Crystal structure of archaeal tyrosyl-tRNA synthetase complexed with tRNA(Tyr) and L-tyrosine | | Descriptor: | MAGNESIUM ION, TYROSINE, Tyrosyl-tRNA synthetase, ... | | Authors: | Kobayashi, T, Nureki, O, Ishitani, R, Tukalo, M, Cusack, S, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion
NAT.STRUCT.BIOL., 10, 2003
|
|
1J2W
 
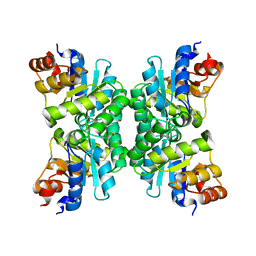 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1KH2
 
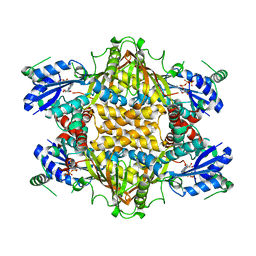 | |
1KH1
 
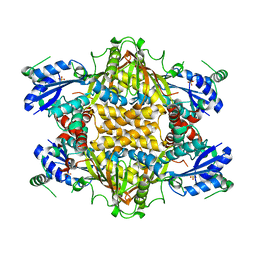 | |
2KHE
 
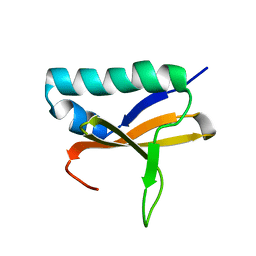 | | Solution Structure of the Bacterial Toxin Rele from Thermus Thermophilus HB8 | | Descriptor: | Toxin-like protein | | Authors: | Suzuki, S, Kawazoe, M, Kaminishi, T, Takemoto, C, Muto, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacterial Toxin Rele from Thermus Thermophilus HB8
To be Published
|
|
1IYW
 
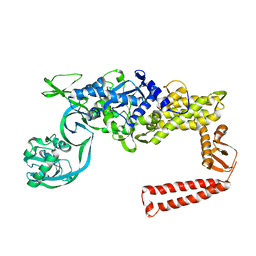 | | Preliminary Structure of Thermus thermophilus Ligand-Free Valyl-tRNA Synthetase | | Descriptor: | Valyl-tRNA Synthetase | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-09-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
1J3L
 
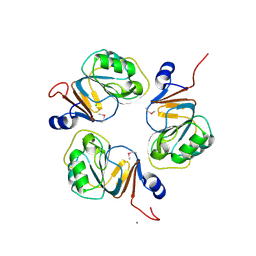 | | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis | | Descriptor: | CHLORIDE ION, Demethylmenaquinone Methyltransferase, MAGNESIUM ION | | Authors: | Rehse, P.H, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-04 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J20
 
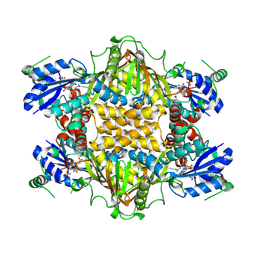 | |
1IUF
 
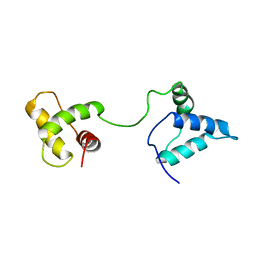 | | LOW RESOLUTION SOLUTION STRUCTURE OF THE TWO DNA-BINDING DOMAINS IN Schizosaccharomyces pombe ABP1 PROTEIN | | Descriptor: | centromere abp1 protein | | Authors: | Kikuchi, J, Iwahara, J, Kigawa, T, Murakami, Y, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure determination of the two DNA-binding domains in the Schizosaccharomyces pombe Abp1 protein by a combination of dipolar coupling and diffusion anisotropy restraints.
J.Biomol.NMR, 22, 2002
|
|
1IUG
 
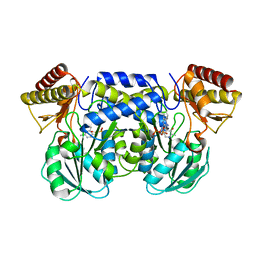 | | The crystal structure of aspartate aminotransferase which belongs to subgroup IV from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, putative aspartate aminotransferase | | Authors: | Katsura, Y, Shirouzu, M, Yamaguchi, H, Ishitani, R, Nureki, O, Kuramitsu, S, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV.
Proteins, 55, 2004
|
|
1IXD
 
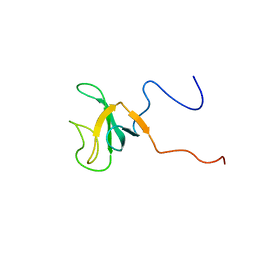 | | Solution structure of the CAP-GLY domain from human cylindromatosis tomour-suppressor CYLD | | Descriptor: | Cylindromatosis tumour-suppressor CYLD | | Authors: | Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma
STRUCTURE, 12, 2004
|
|
1IYF
 
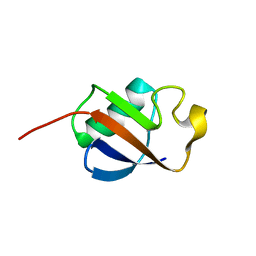 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
1IZ0
 
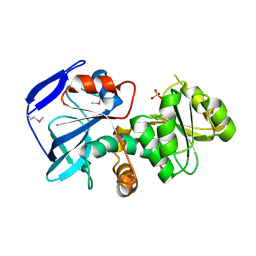 | |
1ILE
 
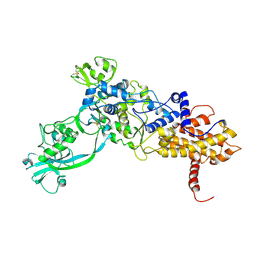 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
1HLV
 
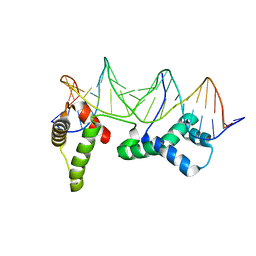 | | CRYSTAL STRUCTURE OF CENP-B(1-129) COMPLEXED WITH THE CENP-B BOX DNA | | Descriptor: | CENP-B BOX DNA, MAJOR CENTROMERE AUTOANTIGEN B | | Authors: | Tanaka, Y, Nureki, O, Kurumizaka, H, Fukai, S, Kawaguchi, S, Ikuta, M, Iwahara, J, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-04 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA.
EMBO J., 20, 2001
|
|
2IDE
 
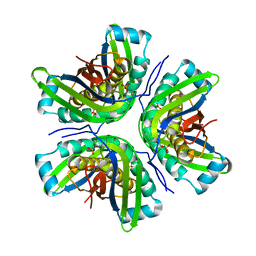 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
2III
 
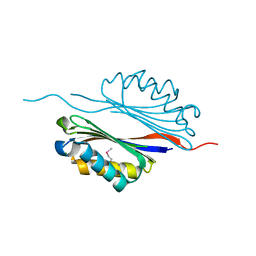 | | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5
To be Published
|
|
