7T7L
 
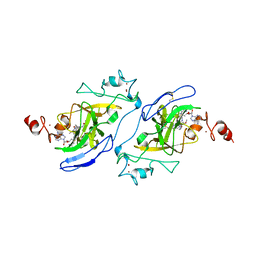 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7T7M
 
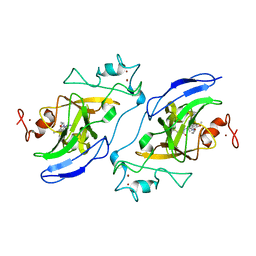 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7X1X
 
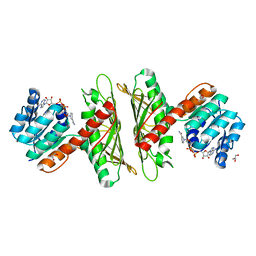 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7X2Y
 
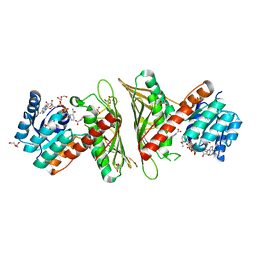 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7WZD
 
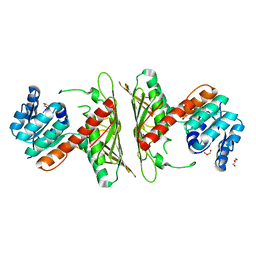 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase from Comamonas testosteroni KF1 | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7DLK
 
 | | Crystal Structure of veratryl alcohol bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2020-11-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dye-decolorizing peroxidase from Bacillus subtilis in complex with veratryl alcohol.
Int.J.Biol.Macromol., 193, 2021
|
|
7E5Q
 
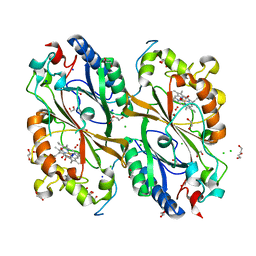 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis at acidic pH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2021-02-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights at acidic pH of dye-decolorizing peroxidase from Bacillus subtilis.
Proteins, 2022
|
|
3IIR
 
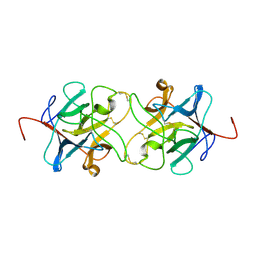 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
1LJX
 
 | | THE STRUCTURE OF D(TPGPCPGPCPA)2 AT 293K: COMPARISON OF THE EFFECT OF SEQUENCE AND TEMPERATURE | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*A)-3', MAGNESIUM ION | | Authors: | Thiyagarajan, S, Satheesh Kumar, P, Rajan, S.S, Gautham, N. | | Deposit date: | 2002-04-23 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of d(TGCGCA)2 at 293 K: comparison of the effects of sequence and temperature.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5G4B
 
 | |
6DGC
 
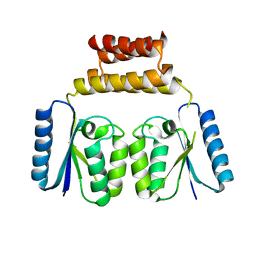 | | Crystal structure of the C-terminal catalytic domain of ISC1926 TnpA, an IS607-like serine recombinase | | Descriptor: | ISC1926 TnpA C-terminal catalytic domain | | Authors: | Hancock, S.P, Kumar, P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
3MGZ
 
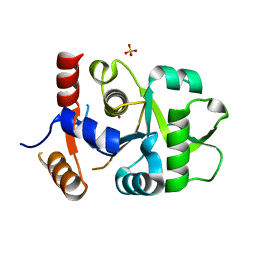 | | Crystal structure of DHBPS domain of bi-functional DHBPS/GTP cyclohydrolase II from Mycobacterium tuberculosis at pH 4.0 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION | | Authors: | Singh, M, Kumar, P, Karthikeyan, S. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
7FHR
 
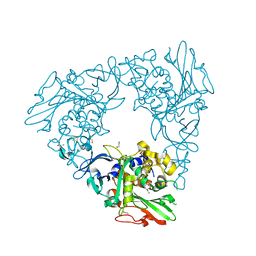 | | Crystal Structure of a Rieske Oxygenase from Cupriavidus metallidurans | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mahto, J.K, Dhankhar, P, Kumar, P. | | Deposit date: | 2021-07-30 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by phthalate dioxygenase from Comamonas testosteroni.
J.Biol.Chem., 297, 2021
|
|
6LLA
 
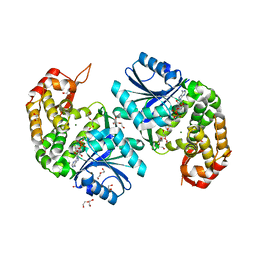 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+ and NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
1I6Q
 
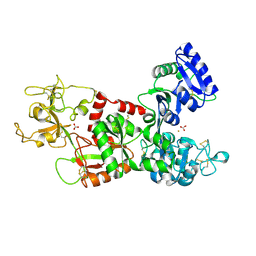 | | Formation of a protein intermediate and its trapping by the simultaneous crystallization process: Crystal structure of an iron-saturated intermediate in the FE3+ binding pathway of camel lactoferrin at 2.7 resolution | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Khan, J.A, Kumar, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2001-03-03 | | Release date: | 2001-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein intermediate trapped by the simultaneous crystallization process. Crystal structure of an iron-saturated intermediate in the Fe3+ binding pathway of camel lactoferrin at 2.7 a resolution.
J.Biol.Chem., 276, 2001
|
|
7V25
 
 | |
7V28
 
 | |
3ZHW
 
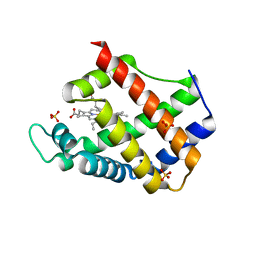 | | X-ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and their Functional Implications | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Mukhi, N, Dhindwal, S, Uppal, S, Kumar, P, Kaur, J, Kundu, S. | | Deposit date: | 2012-12-30 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | X-Ray Crystallographic Structural Characteristics of Arabidopsis Hemoglobin I and Their Functional Implications
Biochim.Biophys.Acta, 1834, 2013
|
|
1DTZ
 
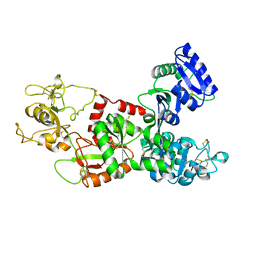 | | STRUCTURE OF CAMEL APO-LACTOFERRIN DEMONSTRATES ITS DUAL ROLE IN SEQUESTERING AND TRANSPORTING FERRIC IONS SIMULTANEOUSLY:CRYSTAL STRUCTURE OF CAMEL APO-LACTOFERRIN AT 2.6A RESOLUTION. | | Descriptor: | APO LACTOFERRIN | | Authors: | Khan, J.A, Kumar, P, Paramasivam, M, Srinivasan, A, Yadav, R.S, Sahani, M.S, Singh, T.P. | | Deposit date: | 2000-01-13 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Camel lactoferrin, a transferrin-cum-lactoferrin: crystal structure of camel apolactoferrin at 2.6 A resolution and structural basis of its dual role.
J.Mol.Biol., 309, 2001
|
|
4AN6
 
 | |
4AN7
 
 | |
5FTW
 
 | | Crystal structure of glutamate O-methyltransferase in complex with S- adenosyl-L-homocysteine (SAH) from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHEMOTAXIS PROTEIN METHYLTRANSFERASE, GLYCEROL, ... | | Authors: | Sharma, R, Dhindwal, S, Batra, M, Aggarwal, M, Kumar, P, Tomar, S. | | Deposit date: | 2016-01-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pentapeptide-Independent Chemotaxis Receptor Methyltransferase (Cher) Reveals Idiosyncratic Structural Determinants for Receptor Recognition.
J.Struct.Biol., 196, 2016
|
|
3ZQ7
 
 | |
4C44
 
 | | Crystal Structure of Truncated Plant Hemoglobin from Arabidopsis thaliana | | Descriptor: | 2-ON-2 HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION | | Authors: | Mukhi, N, Dhindwal, S, Kumar, P, Kaur, J, Kundu, S. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-Ray Crystallographic Structural Characteristics of Truncated Hemoglobin from Arabidopsis Thaliana
To be Published
|
|
4U36
 
 | | Crystal structure of a seed lectin from Vatairea macrocarpa complexed with Tn-antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
