1H7M
 
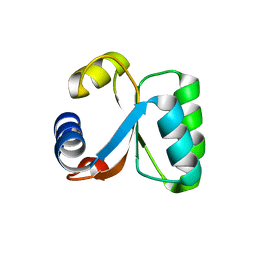 | | Ribosomal Protein L30e from Thermococcus celer | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Chen, Y.W, Wong, K.B. | | Deposit date: | 2001-07-09 | | Release date: | 2003-04-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Ribosomal Protein L30E from the Extreme Thermophile Thermocccus Celer: Thermal Stability and RNA Binding
Biochemistry, 42, 2003
|
|
1GO1
 
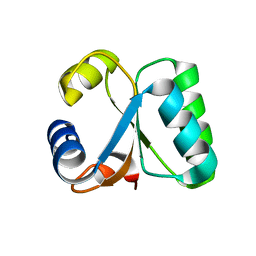 | |
2R0F
 
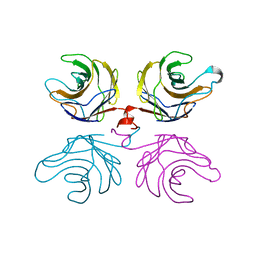 | | Ligand free structure of fungal lectin CGL3 | | Descriptor: | CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
2R0H
 
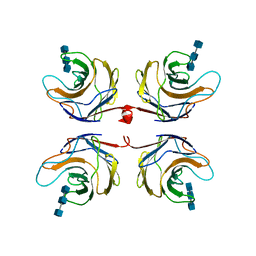 | | Fungal lectin CGL3 in complex with chitotriose (chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-20 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
1JJ2
 
 | | Fully Refined Crystal Structure of the Haloarcula marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The kink-turn: a new RNA secondary structure motif.
EMBO J., 20, 2001
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDF
 
 | |
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECV
 
 | | Bovine Fab 2F12 | | Descriptor: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1SKM
 
 | | HhaI methyltransferase in complex with DNA containing an abasic south carbocyclic sugar at its target site | | Descriptor: | 5'-D(*T*GP*TP*CP*AP*GP*(HCX)P*GP*CP*AP*TP*GP*G)-3', 5'-D(*TP*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', Modification methylase HhaI, ... | | Authors: | Horton, J.R, Ratner, G, Banavali, N, Huang, N, Marquez, V.E, MacKerell, A.D, Cheng, X. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase
Nucleic Acids Res., 32, 2004
|
|
8GC8
 
 | |
8GC7
 
 | |
6PU7
 
 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
6WJY
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4-A | | Descriptor: | 3-chloro-N-(3-{(2S)-1-[(4-fluorophenyl)amino]-1-oxopropan-2-yl}bicyclo[1.1.1]pentan-1-yl)benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A, Neumann, L. | | Deposit date: | 2020-04-14 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of Potent and Orally Available Bicyclo[1.1.1]pentane-Derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7RRD
 
 | | IDO1 IN COMPLEX WITH COMPOUND S-1 | | Descriptor: | 3-[4-(1H-benzimidazol-2-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Oxetane promise delivered: discovery of long acting IDO1 inhibitors suitable for Q3W oral or parenteral dosing
To Be Published
|
|
7SJS
 
 | |
7RRC
 
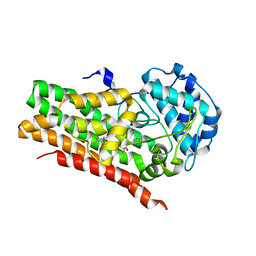 | | IDO1 IN COMPLEX WITH COMPOUND 14 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-fluorophenyl)-3-{4-[4-(hydroxymethyl)-6-(trifluoromethyl)pyridin-3-yl]phenyl}oxetane-3-carboxamide | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRB
 
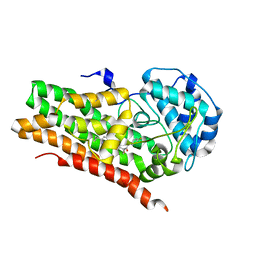 | | IDO1 IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 3-[4-(6-cyclopropylpyridin-3-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
6X5Y
 
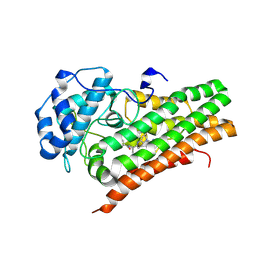 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
2MMM
 
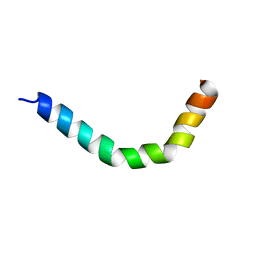 | |
6G0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SIRT7 peptide (K272ac/K275ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, NAD-dependent protein deacetylase sirtuin-7 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0R
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated POLR2A peptide (K775ac/K778ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DNA-directed RNA polymerase II subunit RPB1 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0Q
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Erythroid transcription factor | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
