7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | 分子名称: | DNA (168-MER), Histone H2A, Histone H2B, ... | | 著者 | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | 登録日 | 2020-08-14 | | 公開日 | 2020-10-21 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
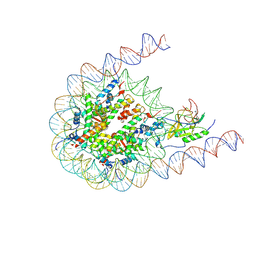 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | 分子名称: | DNA (168-MER), Histone H2A, Histone H2B, ... | | 著者 | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | 登録日 | 2020-08-14 | | 公開日 | 2020-10-21 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | 分子名称: | DNA (168-MER), Histone H2A, Histone H2B, ... | | 著者 | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | 登録日 | 2020-08-14 | | 公開日 | 2020-10-21 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
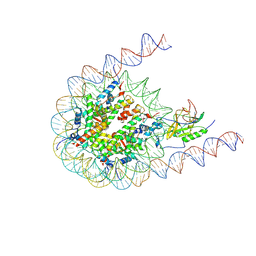 | | Native NSD3 bound to 187-bp nucleosome | | 分子名称: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | 著者 | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | 登録日 | 2020-08-14 | | 公開日 | 2020-10-21 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
2L7Y
 
 | |
5ZWK
 
 | | Crystal structure of Human liver fructose-1,6-bisphoaphatase complex with fructose-1,6-bisphophate and AMP | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | 著者 | Yunyuan, H, Zeyuan, G, Junjie, Y, Ping, Y, Jian, W. | | 登録日 | 2018-05-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Location of FBPase catalytic metal binding site: a combined experimental and theoretical study
To Be Published
|
|
8IJ5
 
 | |
7DV8
 
 | |
6KIZ
 
 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIX
 
 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
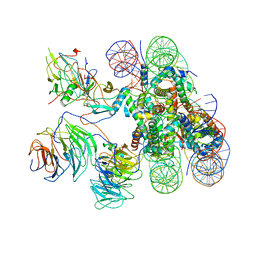 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
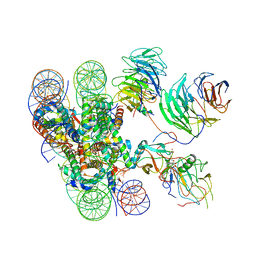 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6J99
 
 | |
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | 著者 | Liu, Z, Liu, S, Yuanzhu, G. | | 登録日 | 2022-03-27 | | 公開日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
6JZE
 
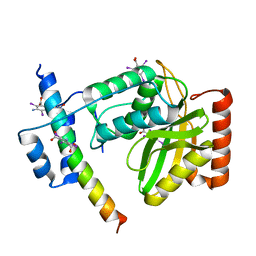 | | Crystal structure of VASH2-SVBP complex with the magic triangle I3C | | 分子名称: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | 著者 | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | 登録日 | 2019-05-01 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-04-05 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
6JZD
 
 | | Crystal structure of peptide-bound VASH2-SVBP complex | | 分子名称: | GLU-GLY-GLU-GLU-TYR, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | 著者 | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | 登録日 | 2019-05-01 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.479 Å) | | 主引用文献 | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
6JZC
 
 | | Structural basis of tubulin detyrosination | | 分子名称: | GLYCEROL, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | 著者 | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | 登録日 | 2019-05-01 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
