1BQT
 
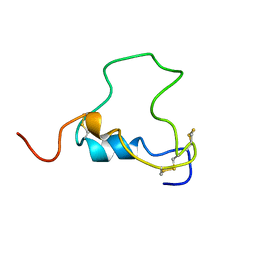 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | 分子名称: | INSULIN-LIKE GROWTH FACTOR-I | | 著者 | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | 登録日 | 1998-08-18 | | 公開日 | 1999-05-18 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
1TTK
 
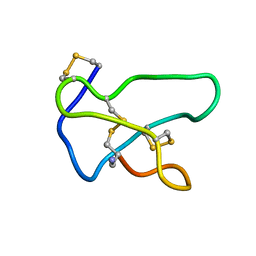 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | 分子名称: | Omega-conotoxin MVIIa | | 著者 | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | 登録日 | 2004-06-22 | | 公開日 | 2004-07-06 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TT3
 
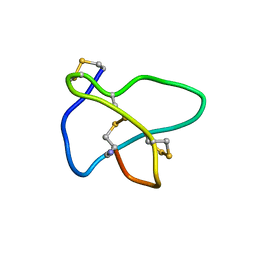 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | 分子名称: | Omega-conotoxin MVIIa | | 著者 | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | 登録日 | 2004-06-21 | | 公開日 | 2004-07-06 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TTL
 
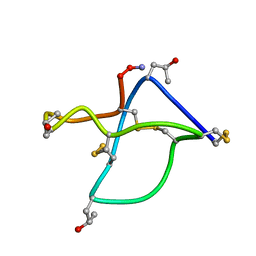 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | 分子名称: | Omega-conotoxin GVIA | | 著者 | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | 登録日 | 2004-06-23 | | 公開日 | 2004-07-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1TR6
 
 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | 分子名称: | Omega-conotoxin GVIA | | 著者 | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | 登録日 | 2004-06-21 | | 公開日 | 2004-07-13 | | 最終更新日 | 2011-10-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
5XRZ
 
 | | Structure of a ssDNA bound to the inner DNA binding site of RAD52 | | 分子名称: | DNA repair protein RAD52 homolog, POTASSIUM ION, ssDNA (40-MER) | | 著者 | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | 登録日 | 2017-06-11 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
5XS0
 
 | | Structure of a ssDNA bound to the outer DNA binding site of RAD52 | | 分子名称: | DNA repair protein RAD52 homolog, ssDNA (5'-D(*CP*CP*CP*CP*CP*C)-3'), ssDNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | 著者 | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | 登録日 | 2017-06-11 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
8I5I
 
 | |
8I5H
 
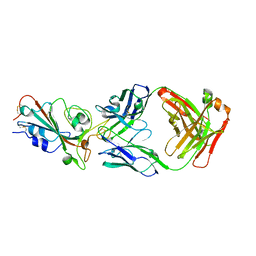 | |
7WN2
 
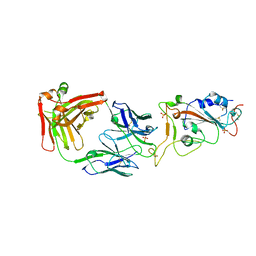 | |
7WNB
 
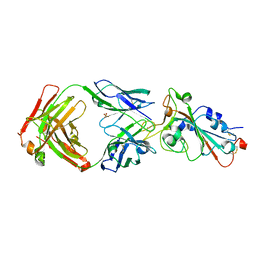 | |
7YOW
 
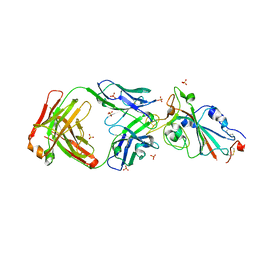 | |
7CLX
 
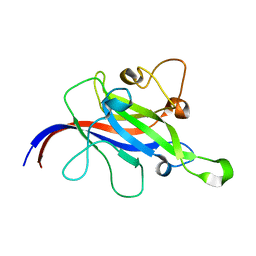 | | Crystal structure of the DOCK8 DHR-1 domain | | 分子名称: | Dedicator of cytokinesis protein 8 | | 著者 | Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S, Fukui, Y, Uruno, T. | | 登録日 | 2020-07-22 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A conserved PI(4,5)P2-binding domain is critical for immune regulatory function of DOCK8.
Life Sci Alliance, 4, 2021
|
|
7CLY
 
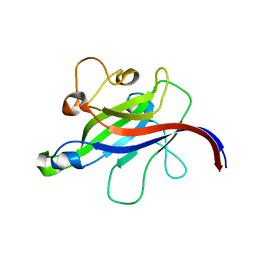 | | Structure of the DOCK8 DHR-1 domain crystallized with di-C8-phosphatidylinositol-(4,5)-bisphosphate | | 分子名称: | Dedicator of cytokinesis protein 8 | | 著者 | Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S, Fukui, Y, Uruno, T. | | 登録日 | 2020-07-22 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.432 Å) | | 主引用文献 | A conserved PI(4,5)P2-binding domain is critical for immune regulatory function of DOCK8.
Life Sci Alliance, 4, 2021
|
|
4A11
 
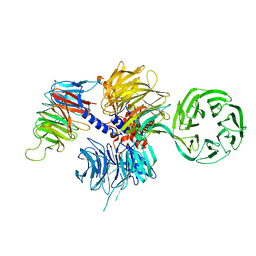 | | Structure of the hsDDB1-hsCSA complex | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | 著者 | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | 登録日 | 2011-09-13 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0K
 
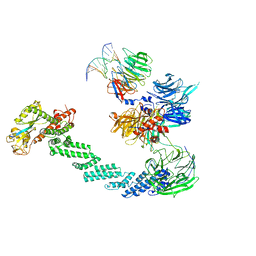 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | 分子名称: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (5.93 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | 分子名称: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (7.4 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
