7YQQ
 
 | | Crystal Structure of Xcc NAMPT and its complex with NMN | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, PHOSPHATE ION, Pre-B cell enhancing factor related protein | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2022-08-08 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure of Xanthomonas campestris pv. campestris Nicotinamide Phosphoribosyltransferase: insights into bacterial NAD+ biosynthesis from the salvage pathway
To Be Published
|
|
7YQP
 
 | | Xcc Nicotinamide Phosphoribosyltransferase | | 分子名称: | Pre-B cell enhancing factor related protein | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2022-08-08 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.088 Å) | | 主引用文献 | Structure of Xanthomonas campestris pv. campestris Nicotinamide Phosphoribosyltransferase: insights into bacterial NAD+ biosynthesis from the salvage pathway
To Be Published
|
|
7YQO
 
 | | Xcc Nicotinamide Phosphoribosyltransferase | | 分子名称: | GLYCEROL, Pre-B cell enhancing factor related protein | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2022-08-08 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure of Xanthomonas campestris pv. campestris Nicotinamide Phosphoribosyltransferase: insights into bacterial NAD+ biosynthesis from the salvage pathway
To Be Published
|
|
7YQR
 
 | | Crystal Structure of Xcc NAMPT and its complex with NAM | | 分子名称: | NICOTINAMIDE, Pre-B cell enhancing factor related protein | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2022-08-08 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Xanthomonas campestris pv. campestris Nicotinamide Phosphoribosyltransferase: insights into bacterial NAD+ biosynthesis from the salvage pathway
To Be Published
|
|
4LAK
 
 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in apo form | | 分子名称: | Uracil-5-carboxylate decarboxylase, ZINC ION | | 著者 | Xu, S, Li, W, Zhu, J, Wang, R, Li, Z, Xu, G.L, Ding, J. | | 登録日 | 2013-06-20 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
8IGZ
 
 | | Xcc NAMPT Quadruple mutant | | 分子名称: | Pre-B cell enhancing factor related protein | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2023-02-21 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7W3R
 
 | | USP34 catalytic domain | | 分子名称: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2021-11-26 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7W3U
 
 | | USP34 catalytic domain in complex with UbPA | | 分子名称: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | 著者 | Xu, G.L, Ming, Z.H. | | 登録日 | 2021-11-26 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
6JV3
 
 | |
6JV5
 
 | |
4LAN
 
 | | Crystal structure of Cordyceps militaris IDCase H195A mutant | | 分子名称: | Uracil-5-carboxylate decarboxylase, ZINC ION | | 著者 | Xu, S, Li, W, Zhu, J, Ding, J. | | 登録日 | 2013-06-20 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAL
 
 | | Crystal structure of Cordyceps militaris IDCase D323A mutant in complex with 5-carboxyl-uracil | | 分子名称: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | 著者 | Xu, S, Li, W, Zhu, J, Ding, J. | | 登録日 | 2013-06-20 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAM
 
 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in complex with 5-carboxyl-uracil | | 分子名称: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | 著者 | Xu, S, Li, W, Zhu, J, Ding, J. | | 登録日 | 2013-06-20 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAO
 
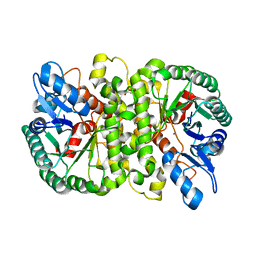 | | Crystal structure of Cordyceps militaris IDCase H195A mutant (Zn) | | 分子名称: | Cordyceps militaris IDCase, DI(HYDROXYETHYL)ETHER, ZINC ION | | 著者 | Xu, S, Li, W, Zhu, J, Ding, J. | | 登録日 | 2013-06-20 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
5EXH
 
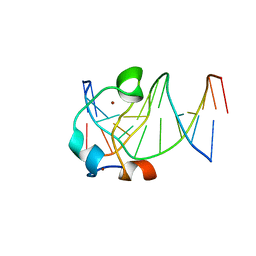 | |
7CY6
 
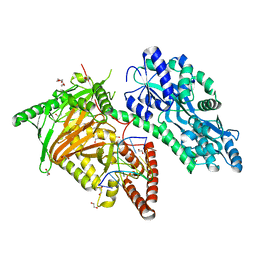 | | Crystal Structure of CMD1 in complex with 5mC-DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY7
 
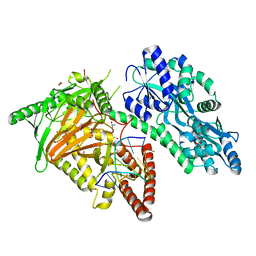 | | Crystal Structure of CMD1 in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY5
 
 | | Crystal Structure of CMD1 in complex with vitamin C | | 分子名称: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY8
 
 | | Crystal Structure of CMD1 in complex with 5mC-DNA and vitamin C | | 分子名称: | 1,2-ETHANEDIOL, ASCORBIC ACID, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY4
 
 | | Crystal Structure of CMD1 in apo form | | 分子名称: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
5ZAT
 
 | |
5ZAS
 
 | |
3UO7
 
 | |
3UOB
 
 | |
4HK6
 
 | |
