1TVP
 
 | |
1TVN
 
 | |
2WHT
 
 | |
2WHS
 
 | | Fluorescent Protein mKeima at pH 3.8 | | 分子名称: | LARGE STOKES SHIFT FLUORESCENT PROTEIN, SULFATE ION | | 著者 | Violot, S, Carpentier, P, Blanchoin, L, Bourgeois, D. | | 登録日 | 2009-05-06 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Reverse Ph-Dependence of Chromophore Protonation Explains the Large Stokes Shift of the Red Fluorescent Protein Mkeima.
J.Am.Chem.Soc., 131, 2009
|
|
2WHU
 
 | |
7O7T
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c in complex with 4-deoxy-L-erythro-5-hexoseulose uronic acid | | 分子名称: | 1,2-ETHANEDIOL, 4-deoxy-L-erythro-hex-5-ulosuronic acid, GLYCEROL, ... | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O84
 
 | | Structure of the PL6 family alginate lyase Pedsa0632 from Pseudopedobacter saltans in complex with substrate | | 分子名称: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.177 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O77
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c | | 分子名称: | GLYCEROL, Poly(Beta-D-mannuronate) lyase | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.321 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O78
 
 | | Structure of the PL6 family chondroitinase B from Pseudopedobacter saltans, Pedsa3807 | | 分子名称: | Polysaccharide lyase from Pseudopedobacter saltans, Pedsa3807 | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O7A
 
 | |
7O79
 
 | | Structure of the PL6 family polysaccharide lyase Pedsa3628 from Pseudopedobacter saltans | | 分子名称: | PHOSPHATE ION, Poly(Beta-D-mannuronate) lyase | | 著者 | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | 登録日 | 2021-04-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7QP1
 
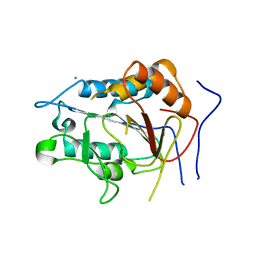 | | Crystal structure of metacaspase from candida glabrata with calcium | | 分子名称: | CALCIUM ION, CHLORIDE ION, Metacaspase-1 | | 著者 | Conchou, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2021-12-30 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol, 5, 2022
|
|
7QP0
 
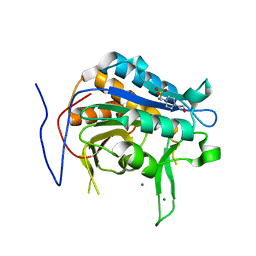 | | Crystal structure of metacaspase from candida glabrata with magnesium | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Metacaspase-1 | | 著者 | Conchou, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2021-12-30 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol, 5, 2022
|
|
7ZU9
 
 | |
4WIN
 
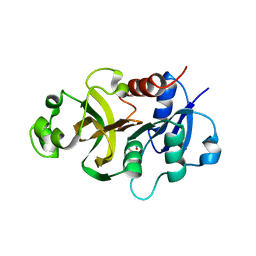 | | Crystal structure of the GATase domain from Plasmodium falciparum GMP synthetase | | 分子名称: | GMP synthetase, NITRATE ION | | 著者 | Ballut, L, Violot, S, Haser, R, Aghajari, N. | | 登録日 | 2014-09-26 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Active site coupling in Plasmodium falciparum GMP synthetase is triggered by domain rotation.
Nat Commun, 6, 2015
|
|
4WIM
 
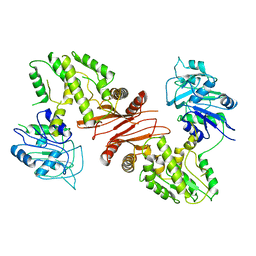 | |
4WIO
 
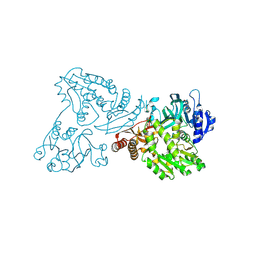 | | Crystal structure of the C89A GMP synthetase inactive mutant from Plasmodium falciparum in complex with glutamine | | 分子名称: | GLUTAMINE, GMP synthetase | | 著者 | Ballut, L, Violot, S, Haser, R, Aghajari, N. | | 登録日 | 2014-09-26 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Active site coupling in Plasmodium falciparum GMP synthetase is triggered by domain rotation.
Nat Commun, 6, 2015
|
|
7P44
 
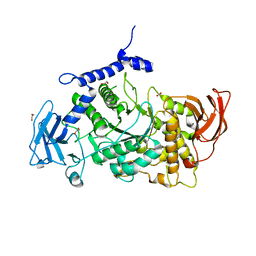 | | Structure of CgGBE in P21212 space group | | 分子名称: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | 著者 | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | 登録日 | 2021-07-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P45
 
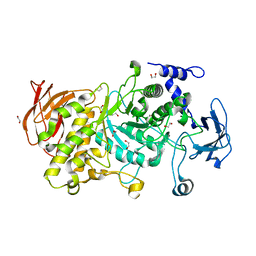 | | Structure of CgGBE in P212121 space group | | 分子名称: | 1,2-ETHANEDIOL, 1,4-alpha-glucan-branching enzyme | | 著者 | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | 登録日 | 2021-07-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
7P43
 
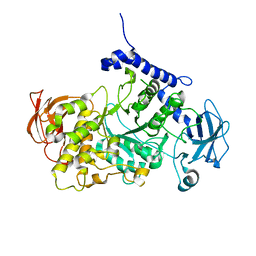 | | Structure of CgGBE in complex with maltotriose | | 分子名称: | 1,4-alpha-glucan-branching enzyme, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | 登録日 | 2021-07-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
6RNH
 
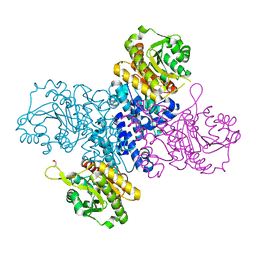 | | Structure of C-terminal truncated Plasmodium falciparum IMP-nucleotidase | | 分子名称: | GLYCEROL, IMP-specific 5'-nucleotidase, putative | | 著者 | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2019-05-08 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RN1
 
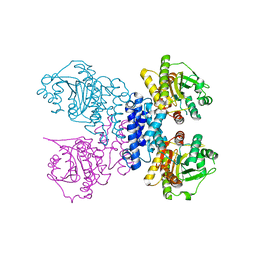 | | Structure of N-terminal truncated Plasmodium falciparum IMP-nucleotidase | | 分子名称: | IMP-specific 5'-nucleotidase, putative | | 著者 | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2019-05-07 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMO
 
 | | Structure of Plasmodium falciparum IMP-nucleotidase | | 分子名称: | IMP-specific 5'-nucleotidase, putative | | 著者 | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2019-05-07 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RME
 
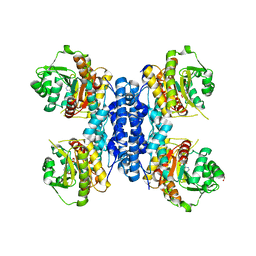 | | Structure of IMP bound Plasmodium falciparum IMP-nucleotidase mutant D172N | | 分子名称: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | 著者 | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2019-05-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMD
 
 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | 著者 | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | 登録日 | 2019-05-06 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
