6IYB
 
 | | Structure of human ORP1 ANK - Rab7 complex | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Oxysterol-binding protein-related protein 1, ... | | 著者 | Tong, J, Im, Y.J. | | 登録日 | 2018-12-14 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.091 Å) | | 主引用文献 | Structural basis of human ORP1-Rab7 interaction for the late-endosome and lysosome targeting.
PLoS ONE, 14, 2019
|
|
4INQ
 
 | | Crystal structure of Osh3 ORD in complex with PI(4)P from Saccharomyces cerevisiae | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Oxysterol-binding protein homolog 3 | | 著者 | Tong, J, Im, Y.J. | | 登録日 | 2013-01-05 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins
Structure, 21, 2013
|
|
4IC4
 
 | |
5YQQ
 
 | |
5YQI
 
 | | Crystal structure of the first StARkin domain of Lam2 | | 分子名称: | Membrane-anchored lipid-binding protein YSP2 | | 著者 | Tong, J, Im, Y.J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YQP
 
 | |
5YS0
 
 | |
5YQR
 
 | | Crystal structure of the PH-like domain of Lam6 | | 分子名称: | Endolysin/Membrane-anchored lipid-binding protein LAM6 fusion protein, NONAETHYLENE GLYCOL | | 著者 | Tong, J, Im, Y.J. | | 登録日 | 2017-11-07 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7DEI
 
 | | Structure of human ORP3 ORD domain in complex with PI(4)P | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Oxysterol-binding protein-related protein 3 | | 著者 | Tong, J, Tan, L, Im, Y.J. | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of human ORP3 ORD reveals conservation of a key function and ligand specificity in OSBP-related proteins.
Plos One, 16, 2021
|
|
7DEJ
 
 | | Structure of human ORP3 ORD in apo-form | | 分子名称: | Oxysterol-binding protein-related protein 3 | | 著者 | Tong, J, Tan, L, Im, Y.J. | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of human ORP3 ORD reveals conservation of a key function and ligand specificity in OSBP-related proteins.
Plos One, 16, 2021
|
|
4IAP
 
 | |
4R0Y
 
 | |
6L1D
 
 | |
6L1M
 
 | |
8XSF
 
 | | SARS-CoV-2 RBD + IMCAS-364 + hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, IMCAS-364 H chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.16 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSI
 
 | | SARS-CoV-2 RBD + IMCAS-364 (Local Refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-364 H chain, IMCAS-364 L chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSE
 
 | | SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 H chain, IMCAS-123 L chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSJ
 
 | | SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSL
 
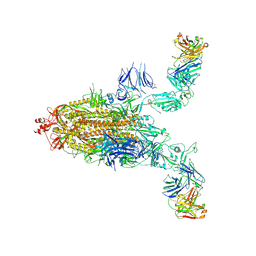 | | SARS-CoV-2 spike + IMCAS-123 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 heavy chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
7V5N
 
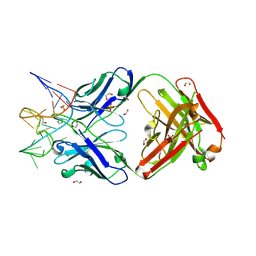 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | 著者 | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | 登録日 | 2021-08-17 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
7F65
 
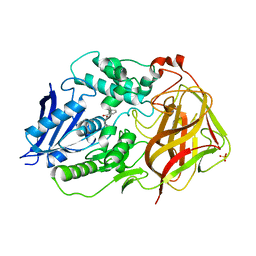 | | Bacetrial Cocaine Esterase with mutations T172R/G173Q/V116K/S117A/A51L, bound to benzoic acid | | 分子名称: | BENZOIC ACID, Cocaine esterase, SULFATE ION | | 著者 | Ouyang, P.F, Zhang, Y, Tong, J. | | 登録日 | 2021-06-24 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5YQJ
 
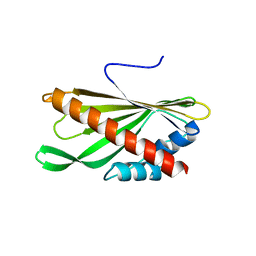 | | Crystal structure of the first StARkin domain of Lam4 | | 分子名称: | Membrane-anchored lipid-binding protein LAM4 | | 著者 | Im, Y.J, Tong, J.S. | | 登録日 | 2017-11-06 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8DSX
 
 | |
7ZL9
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | 著者 | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | 登録日 | 2022-04-14 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZLY
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | 著者 | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | 登録日 | 2022-04-17 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
