2Y2Y
 
 | | Oxidised form of E. coli CsgC | | 分子名称: | ACETATE ION, CURLI PRODUCTION PROTEIN CSGC | | 著者 | Taylor, J.D, Salgado, P.S, Constable, S.C, Cota, E, Mathews, S.J. | | 登録日 | 2010-12-16 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Atomic Resolution Insights Into Curli Fiber Biogenesis.
Structure, 19, 2011
|
|
2Y2T
 
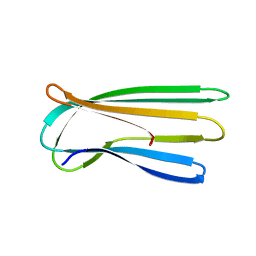 | |
5COM
 
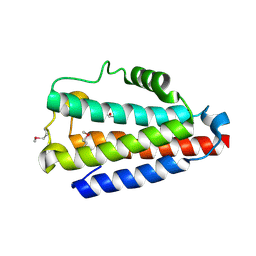 | | Crystal structure of Uncharacterized Protein Q187F5 from Clostridium difficile 630 | | 分子名称: | D(-)-TARTARIC ACID, Putative conjugative transposon protein Tn1549-like, CTn5-Orf2, ... | | 著者 | Taylor, J.D, Taylor, G, Matthews, S.J. | | 登録日 | 2015-07-20 | | 公開日 | 2016-02-03 | | 最終更新日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5CIV
 
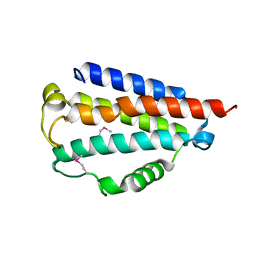 | |
5COF
 
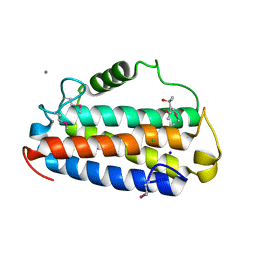 | | Crystal structure of Uncharacterised protein Q1R1X2 from Escherichia coli UTI89 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Taylor, J.D, Hare, S, Matthews, S.J. | | 登録日 | 2015-07-20 | | 公開日 | 2016-02-03 | | 最終更新日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5COG
 
 | | Crystal structure of Yeast IRC4 | | 分子名称: | CHLORIDE ION, IRC4, PHOSPHATE ION, ... | | 著者 | Taylor, J.D, Matthews, S.J. | | 登録日 | 2015-07-20 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.613 Å) | | 主引用文献 | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5CQV
 
 | | Crystal structure of uncharacterized protein Q8DWV2 from Streptococcus agalactiae | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Uncharacterized protein | | 著者 | Taylor, J.D, Hare, S, Matthews, S.J. | | 登録日 | 2015-07-22 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
2JYQ
 
 | | NMR structure of the apo v-Src SH2 domain | | 分子名称: | Tyrosine-protein kinase transforming protein Src | | 著者 | Taylor, J.D, Ababou, A, Williams, M.A, Ladbury, J.E. | | 登録日 | 2007-12-17 | | 公開日 | 2008-06-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, dynamics, and binding thermodynamics of the v-Src SH2 domain: Implications for drug design
Proteins, 73, 2008
|
|
2N59
 
 | | Solution Structure of R. palustris CsgH | | 分子名称: | Putative uncharacterized protein CsgH | | 著者 | Hawthorne, W.J, Taylor, J.D, Escalera-Maurer, A, Lambert, S, Koch, M, Scull, N, Sefer, L, Xu, Y, Matthews, S.J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-05-11 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones.
Sci Rep, 6, 2016
|
|
2XSK
 
 | | E. coli curli protein CsgC - SeCys | | 分子名称: | ACETATE ION, CSGC | | 著者 | Salgado, P.S, Taylor, J.D, Cota, E, Matthews, S.J. | | 登録日 | 2010-09-29 | | 公開日 | 2010-12-29 | | 最終更新日 | 2014-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Extending the Usability of the Phasing Power of Diselenide Bonds: Secys Sad Phasing of Csgc Using a Non-Auxotrophic Strain.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5KH5
 
 | |
5KH4
 
 | |
5KH2
 
 | |
2Y7O
 
 | |
2Y7N
 
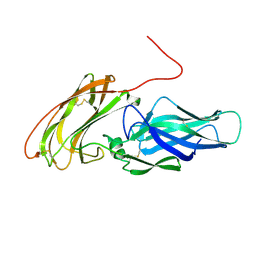 | |
2YLH
 
 | |
2Y7L
 
 | |
2X12
 
 | |
2Y7M
 
 | |
2KUB
 
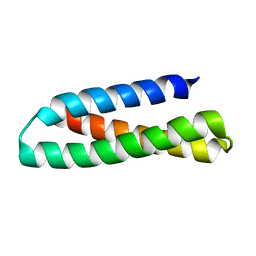 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | 分子名称: | Fimbriae-associated protein Fap1 | | 著者 | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | 登録日 | 2010-02-17 | | 公開日 | 2010-07-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
4G34
 
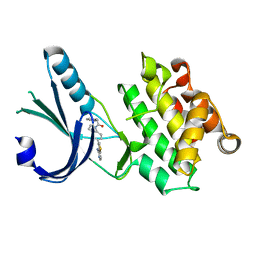 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | 分子名称: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Gampe, R.T, Axten, J.M. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4G31
 
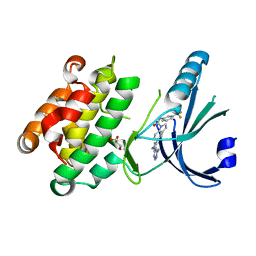 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | 分子名称: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | 著者 | Gampe, R.T, Axten, J.M. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
4Z2A
 
 | | Crystal structure of unglycosylated apo human furin @1.89A | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Furin, ... | | 著者 | Gampe, R.T, Pearce, K, Reid, R. | | 登録日 | 2015-03-29 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | BacMam production and crystal structure of nonglycosylated apo human furin at 1.89A resolution
Acta Crystallogr.,Sect.F, 2019
|
|
4Z2B
 
 | | The structure of human PDE12 residues 161-609 in complex with GSK3036342A | | 分子名称: | 1,2-ETHANEDIOL, 2',5'-phosphodiesterase 12, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | 登録日 | 2015-03-29 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4Z0V
 
 | | The structure of human PDE12 residues 161-609 | | 分子名称: | 2',5'-phosphodiesterase 12, GLYCEROL, MAGNESIUM ION | | 著者 | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | 登録日 | 2015-03-26 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
