7TXT
 
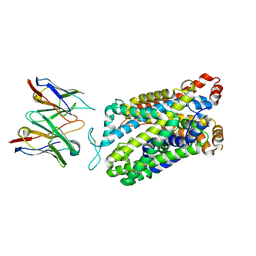 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | 分子名称: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | 著者 | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | 登録日 | 2022-02-09 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
5VBS
 
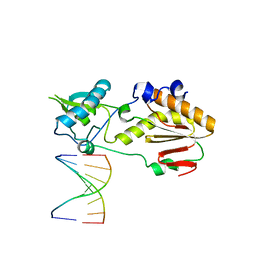 | | Structural basis for a six letter alphabet including GATCKX | | 分子名称: | DNA (5'-D(*CP*TP*TP*AP*TP*(DX)P*(DX)P*T)-3'), DNA (5'-D(P*AP*(93D)P*(93D)P*AP*TP*AP*AP*G)-3'), reverse transcriptase catalytic fragment | | 著者 | Singh, I, Georgiadis, M.M. | | 登録日 | 2017-03-30 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.749 Å) | | 主引用文献 | Structure and Biophysics for a Six Letter DNA Alphabet that Includes Imidazo[1,2-a]-1,3,5-triazine-2(8H)-4(3H)-dione (X) and 2,4-Diaminopyrimidine (K).
ACS Synth Biol, 6, 2017
|
|
5W6Q
 
 | |
5W6K
 
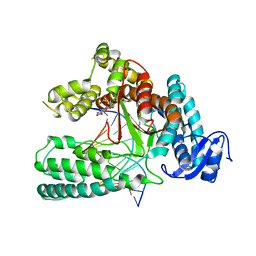 | | Structure of mutant Taq Polymerase incorporating unnatural base pairs Z:P | | 分子名称: | (1R)-1-[6-amino-5-(dihydroxyamino)-2-hydroxypyridin-3-yl]-1,4-anhydro-2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-erythro-pentitol, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA (5'-D(P*(1WA)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | 著者 | Singh, I, Georgiadis, M.M. | | 登録日 | 2017-06-16 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.339 Å) | | 主引用文献 | Snapshots of an evolved DNA polymerase pre- and post-incorporation of an unnatural nucleotide.
Nucleic Acids Res., 46, 2018
|
|
4M94
 
 | |
4M95
 
 | |
6LTQ
 
 | | Crystal structure of pyrrolidone carboxyl peptidase from thermophilic keratin degrading bacterium Fervidobacterium islandicum AW-1 (FiPcp) | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL, Pyroglutamyl-peptidase I | | 著者 | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | 登録日 | 2020-01-23 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of oxidized pyrrolidone carboxypeptidase from Fervidobacterium islandicum AW-1 reveals unique structural features for thermostability and keratinolysis.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7D2N
 
 | |
7D2M
 
 | |
7D28
 
 | |
7D2Q
 
 | |
7D2P
 
 | |
6DPX
 
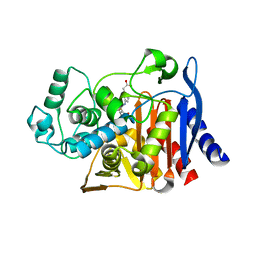 | |
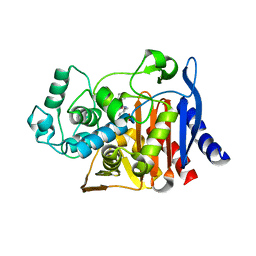
6DPZ
 
 | |
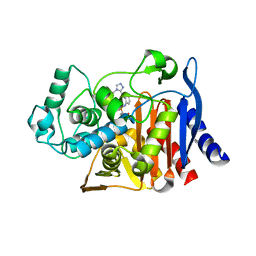
6DPT
 
 | |
8DIC
 
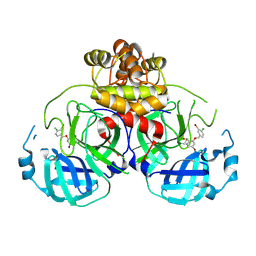 | |
8DIB
 
 | |
8DIG
 
 | |
8DIF
 
 | |
8DII
 
 | |
8DIE
 
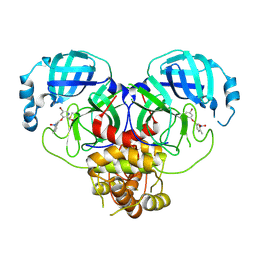 | |
8DIH
 
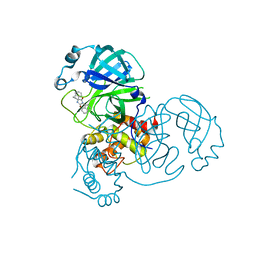 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | 分子名称: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | 著者 | Singh, I, Shoichet, B.K. | | 登録日 | 2022-06-29 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DID
 
 | |
6DPY
 
 | |
6B1R
 
 | |
