8AJR
 
 | |
8ALQ
 
 | |
6HD2
 
 | |
2N8Q
 
 | |
3IT3
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase D261A mutant complexed with substrate 3'-AMP | | 分子名称: | Acid phosphatase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | 著者 | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT1
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with L(+)-tartrate | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, ACETATE ION, Acid phosphatase, ... | | 著者 | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.691 Å) | | 主引用文献 | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT0
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with phosphate | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Acid phosphatase, PENTAETHYLENE GLYCOL, ... | | 著者 | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3IT2
 
 | | Crystal structure of ligand-free Francisella tularensis histidine acid phosphatase | | 分子名称: | ACETATE ION, Acid phosphatase | | 著者 | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | 登録日 | 2009-08-27 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.838 Å) | | 主引用文献 | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3PCT
 
 | |
3OCX
 
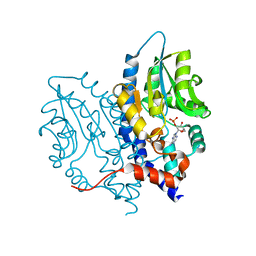 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 2'-AMP | | 分子名称: | ADENOSINE-2'-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OCW
 
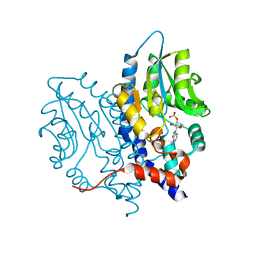 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 3'-AMP | | 分子名称: | Lipoprotein E, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OCZ
 
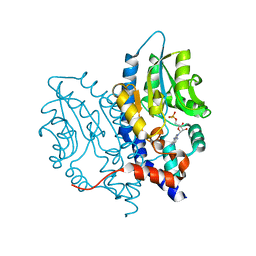 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase Complexed with the inhibitor adenosine 5-O-thiomonophosphate | | 分子名称: | ADENOSINE -5'-THIO-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural basis of the inhibition of class C acid phosphatases by adenosine 5'-phosphorothioate.
Febs J., 278, 2011
|
|
3SF0
 
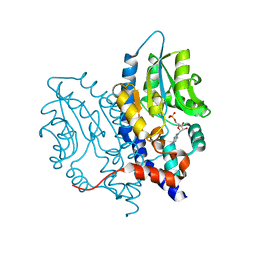 | |
3OCV
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase mutant D66N complexed with 5'-AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.551 Å) | | 主引用文献 | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OCU
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase mutant D66N complexed with NMN | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OCY
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase Complexed with inorganic phosphate | | 分子名称: | Lipoprotein E, MAGNESIUM ION, PHOSPHATE ION | | 著者 | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | 登録日 | 2010-08-10 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
4NMA
 
 | |
4NME
 
 | |
4NMF
 
 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA inactivated by N-propargylglycine and complexed with menadione bisulfite | | 分子名称: | (2R)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, (2S)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, 1,2-ETHANEDIOL, ... | | 著者 | Singh, H, Tanner, J.J. | | 登録日 | 2013-11-14 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NMB
 
 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA in complex with L-lactate | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Singh, H, Almo, S.C, Tanner, J.J. | | 登録日 | 2013-11-14 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NMD
 
 | |
4NM9
 
 | |
4NMC
 
 | |
4DSG
 
 | | Crystal Structure of oxidized UDP-Galactopyranose mutase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | 著者 | Singh, H, Dhatwalia, R, Tanner, J.J. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.249 Å) | | 主引用文献 | Crystal Structures of Trypanosoma cruzi UDP-Galactopyranose Mutase Implicate Flexibility of the Histidine Loop in Enzyme Activation.
Biochemistry, 51, 2012
|
|
4ZTB
 
 | | Crystal structure of nsP2 protease from Chikungunya virus in P212121 space group at 2.59 A (4molecules/ASU). | | 分子名称: | GLYCEROL, Protease nsP2 | | 著者 | Narwal, M, Pratap, S, Singh, H, Kumar, P, Tomar, S. | | 登録日 | 2015-05-14 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site.
Int.J.Biol.Macromol., 116, 2018
|
|
