1SKY
 
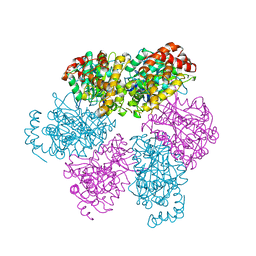 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | 分子名称: | F1-ATPASE, SULFATE ION | | 著者 | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | 登録日 | 1997-02-26 | | 公開日 | 1998-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
6OHD
 
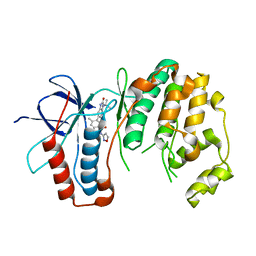 | | P38 in complex with T-3220137 | | 分子名称: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | 著者 | Lane, W, Saikatendu, K. | | 登録日 | 2019-04-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
2RNK
 
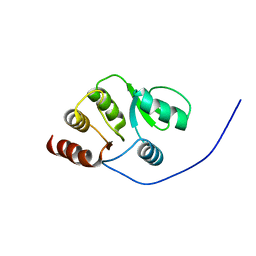 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-11 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
3WKX
 
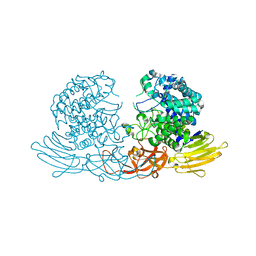 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum arabinose complex form | | 分子名称: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | 著者 | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | 登録日 | 2013-11-01 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
6EBW
 
 | | hALK in complex with compound 9 (6-(((1S)-1-(5-Fluoropyridin-2-yl)ethyl)amino)-1-(3-methyl-1H-pyrazol-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl)(morpholin-4-yl)methanone | | 分子名称: | ALK tyrosine kinase receptor, [6-{[(1S)-1-(5-fluoropyridin-2-yl)ethyl]amino}-1-(5-methyl-1H-pyrazol-3-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl](morpholin-4-yl)methanone | | 著者 | Lane, W, Saikatendu, K. | | 登録日 | 2018-08-07 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.455 Å) | | 主引用文献 | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6E0R
 
 | | hALK in complex with compound 7 N-((1S)-1-(5-fluoropyridin-2-yl)ethyl)-1-(5-methyl-1H-pyrazol-3-yl)-3-(oxetan-3-ylsulfonyl)-1H-pyrrolo[2,3-b]pyridin-6-amine | | 分子名称: | ALK tyrosine kinase receptor, N-[(1S)-1-(5-fluoropyridin-2-yl)ethyl]-1-(5-methyl-1H-pyrazol-3-yl)-3-[(oxetan-3-yl)sulfonyl]-1H-pyrrolo[2,3-b]pyridin-6-amine | | 著者 | Lane, W, Saikatendu, K. | | 登録日 | 2018-07-06 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6EDL
 
 | | hALK in complex with compound 1 (S)-N-(1-(2,4-difluorophenyl)ethyl)-3-(3-methyl-1H-pyrazol-5-yl)imidazo[1,2-b]pyridazin-6-amine | | 分子名称: | ALK tyrosine kinase receptor, N-[(1S)-1-(2,4-difluorophenyl)ethyl]-3-(5-methyl-1H-pyrazol-3-yl)imidazo[1,2-b]pyridazin-6-amine | | 著者 | Lane, W, Saikatendu, K. | | 登録日 | 2018-08-09 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.799 Å) | | 主引用文献 | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
3WKW
 
 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum ligand free form | | 分子名称: | Non-reducing end beta-L-arabinofuranosidase | | 著者 | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | 登録日 | 2013-11-01 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
1WLP
 
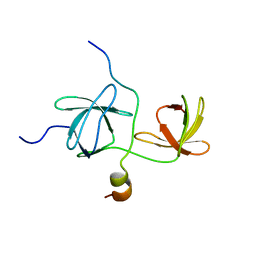 | | Solution Structure Of The P22Phox-P47Phox Complex | | 分子名称: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | 著者 | Ogura, K, Torikai, S, Saikawa, K, Yuzawa, S, Sumimoto, H, Inagaki, F. | | 登録日 | 2004-06-29 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide
J.Biol.Chem., 281, 2006
|
|
2JZE
 
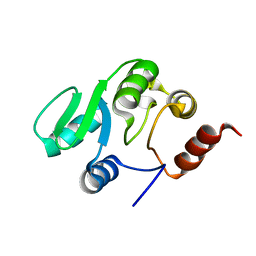 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZD
 
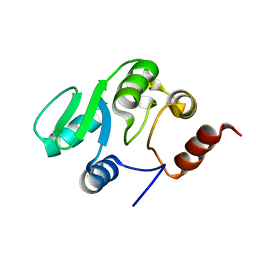 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2OG3
 
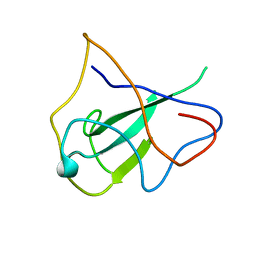 | | structure of the rna binding domain of n protein from SARS coronavirus in cubic crystal form | | 分子名称: | Nucleocapsid protein | | 著者 | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | 登録日 | 2007-01-05 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OFZ
 
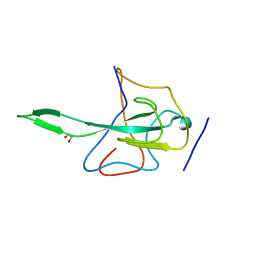 | | Ultrahigh Resolution Crystal Structure of RNA Binding Domain of SARS Nucleopcapsid (N Protein) at 1.1 Angstrom Resolution in Monoclinic Form. | | 分子名称: | 1,2-ETHANEDIOL, Nucleocapsid protein | | 著者 | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | 登録日 | 2007-01-04 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OZK
 
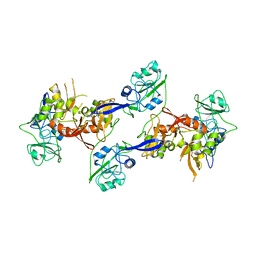 | | Structure of an N-Terminal Truncated Form of Nendou (NSP15) From SARS-CORONAVIRUS | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | 登録日 | 2007-02-26 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch.
J.Virol., 81, 2007
|
|
3LNL
 
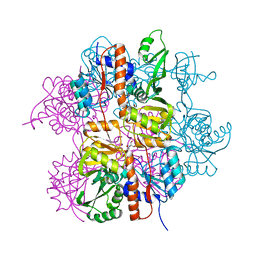 | | Crystal structure of Staphylococcus aureus protein SA1388 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UPF0135 protein SA1388, ZINC ION | | 著者 | Singh, K.S, Chruszcz, M, Zhang, X, Minor, W, Zhang, H. | | 登録日 | 2010-02-02 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a Conserved Hypothetical Protein Sa1388 from S. aureus Reveals a Capped Hexameric Toroid with Two Pii Domain Lids and a Dinuclear Metal Center.
Bmc Struct.Biol., 6, 2006
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | 分子名称: | NSP3 | | 著者 | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2006-09-15 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
4P58
 
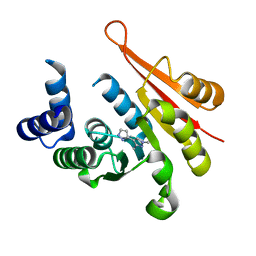 | | Crystal structure of mouse comt bound to an inhibitor | | 分子名称: | 1',3'-dimethyl-1H,1'H-3,4'-bipyrazole, Catechol O-methyltransferase | | 著者 | Lanier, M. | | 登録日 | 2014-03-15 | | 公開日 | 2014-06-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | A fragment-based approach to identifying S-adenosyl-l-methionine -competitive inhibitors of catechol O-methyl transferase (COMT).
J.Med.Chem., 57, 2014
|
|
7BZL
 
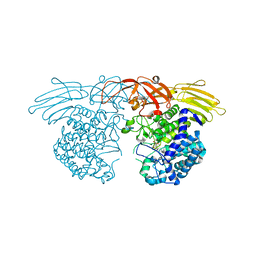 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | 分子名称: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | 登録日 | 2020-04-28 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2HSX
 
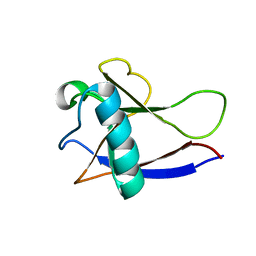 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | 分子名称: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | 著者 | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2006-07-24 | | 公開日 | 2007-02-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | 分子名称: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-11-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4S
 
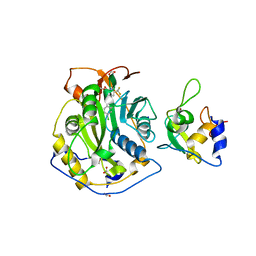 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | 分子名称: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-11-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
2GDT
 
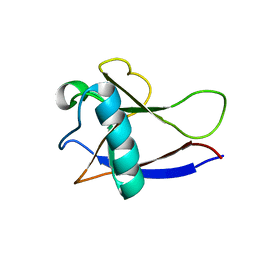 | | NMR Structure of the nonstructural protein 1 (nsp1) from the SARS coronavirus | | 分子名称: | Leader protein; p65 homolog; NSP1 (EC 3.4.22.-) | | 著者 | Almeida, M.S, Herrmann, T, Geralt, M, Johnson, M.A, Saikatendu, K, Joseph, J, Subramanian, R.C, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2006-03-17 | | 公開日 | 2007-02-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2KAF
 
 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | 分子名称: | Non-structural protein 3 | | 著者 | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2008-11-05 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2NYD
 
 | |
