6EUD
 
 | |
7ACO
 
 | |
7ACL
 
 | |
7ACM
 
 | |
5T73
 
 | |
5ORF
 
 | | Structure of ovine serum albumin in P1 space group | | 分子名称: | DI(HYDROXYETHYL)ETHER, PROLINE, Serum albumin, ... | | 著者 | Talaj, J.A, Bujacz, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | 登録日 | 2017-08-16 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5ORI
 
 | | Structure of caprine serum albumin in orthorhombic crystal system | | 分子名称: | Albumin | | 著者 | Bujacz, A, Talaj, J.A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | 登録日 | 2017-08-16 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6YXQ
 
 | | Crystal structure of a DNA repair complex ASCC3-ASCC2 | | 分子名称: | Activating signal cointegrator 1 complex subunit 2, Activating signal cointegrator 1 complex subunit 3 | | 著者 | Jia, J, Absmeier, E, Holton, N, Bohnsack, K.E, Pietrzyk-Brzezinska, A.J, Bohnsack, M.T, Wahl, M.C. | | 登録日 | 2020-05-03 | | 公開日 | 2020-09-30 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.
Nat Commun, 11, 2020
|
|
6ZUP
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-(-)-3-phenyllactic acid | | 分子名称: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, Aminotransferase, MAGNESIUM ION, ... | | 著者 | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | 登録日 | 2020-07-23 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZVG
 
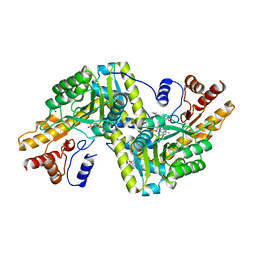 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-indole-3-lactic acid | | 分子名称: | 3-(INDOL-3-YL) LACTATE, Aminotransferase, MAGNESIUM ION, ... | | 著者 | Rum, J, Rutkiewicz, M, Pruska, A, Bujacz, A, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | 登録日 | 2020-07-24 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUR
 
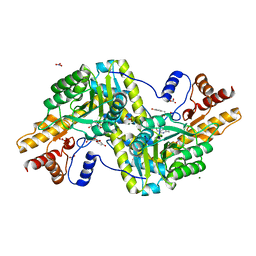 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-p-hydroxyphenyllactic acid | | 分子名称: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, Aminotransferase, MAGNESIUM ION, ... | | 著者 | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | 登録日 | 2020-07-23 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
8QEV
 
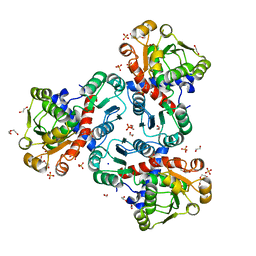 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | 著者 | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | 登録日 | 2023-09-01 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
7ZP0
 
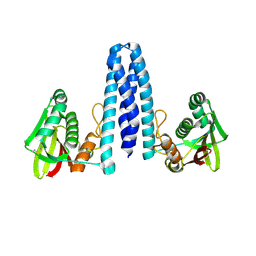 | |
5EUV
 
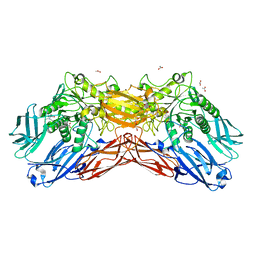 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | 著者 | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | 登録日 | 2015-11-19 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8QEU
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with ornithine | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-ornithine, ... | | 著者 | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | 登録日 | 2023-09-01 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
5OSW
 
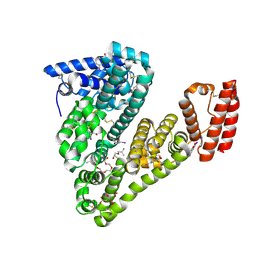 | | Structure of caprine serum albumin in complex with 3,5-diiodosalicylic acid | | 分子名称: | 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, Albumin, ... | | 著者 | Talaj, J.A, Bujacz, A, Bujacz, G. | | 登録日 | 2017-08-18 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OTB
 
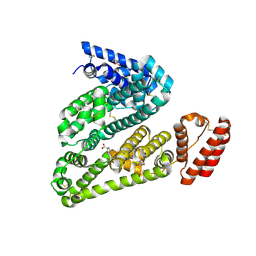 | | Structure of caprine serum albumin in P1 space group | | 分子名称: | Albumin, DI(HYDROXYETHYL)ETHER, PROLINE, ... | | 著者 | Talaj, J.A, Bujacz, A, Bujacz, G. | | 登録日 | 2017-08-21 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LDR
 
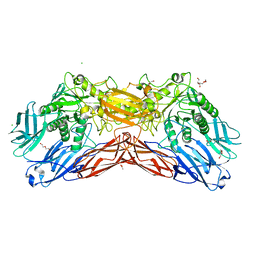 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain in complex with galactose | | 分子名称: | ACETATE ION, Beta-D-galactosidase, CHLORIDE ION, ... | | 著者 | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | 登録日 | 2016-06-27 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6T3V
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - malic acid | | 分子名称: | (2S)-2-hydroxybutanedioic acid, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Rutkiewicz, M, Bujacz, A, Rum, J, Bujacz, G. | | 登録日 | 2019-10-11 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials, 14, 2021
|
|
6ETZ
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB | | 分子名称: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | 著者 | Rutkiewicz-Krotewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2017-10-27 | | 公開日 | 2018-01-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In Situ Random Microseeding and Streak Seeding Used for Growth of Crystals of Cold-Adapted Beta-D-Galactosidases: Crystal Structure of BetaDG from Arthrobacter sp. 32cB
Crystals, 8, 2018
|
|
6H1P
 
 | |
