7NLC
 
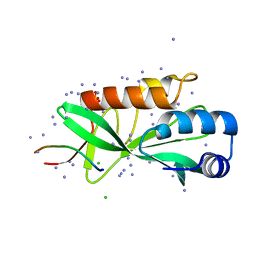 | | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide | | 分子名称: | AMMONIUM ION, CHLORIDE ION, Protein ORF3, ... | | 著者 | Moschidi, D, Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-02-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
To Be Published
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | 分子名称: | Design construct XAA | | 著者 | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | 登録日 | 2019-02-16 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0C
 
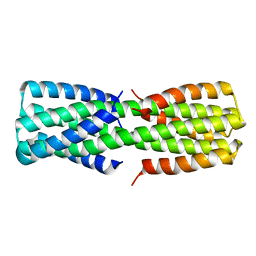 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | 分子名称: | Design construct XAA_GVDQ mutant M4L | | 著者 | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | 登録日 | 2019-02-15 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7P51
 
 | |
8AEB
 
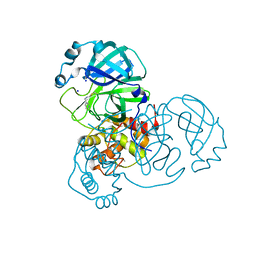 | | SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(pyridin-3-ylmethyl)thioformamide, ... | | 著者 | Hanoulle, X, Charton, J, Deprez, B. | | 登録日 | 2022-07-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
6NY8
 
 | |
6NYE
 
 | |
6NZ1
 
 | |
6NXM
 
 | |
6NZ3
 
 | |
6NX2
 
 | |
6NYI
 
 | |
6NYK
 
 | |
6NPR
 
 | | Crystal structure of H-2Dd with C84-C139 disulfide in complex with gp120 derived peptide P18-I10 | | 分子名称: | ARG-GLY-PRO-GLY-ARG-ALA-PHE-VAL-THR-ILE, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Toor, J, McShan, A.C, Tripathi, S.M, Sgourakis, N.G. | | 登録日 | 2019-01-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7NTS
 
 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | 著者 | Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-03-10 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.477 Å) | | 主引用文献 | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NTQ
 
 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.495 Å) | | 主引用文献 | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
