6Q60
 
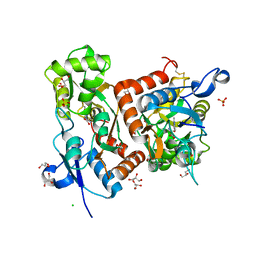 | | Structure of GluA2 ligand-binding domain (S1S2J) in complex with the agonist (S)-2-Amino-3-(2-methyl-5-hydroxy-2H-1,2,3-triazol-4-yl)propanoic acid at 1.55 A resolution | | 分子名称: | (2~{S})-2-azanyl-3-(2-methyl-5-oxidanyl-1,2,3-triazol-4-yl)propanoic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Moellerud, S, Temperini, P, Kastrup, J.S. | | 登録日 | 2018-12-10 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Use of the 4-Hydroxytriazole Moiety as a Bioisosteric Tool in the Development of Ionotropic Glutamate Receptor Ligands.
J.Med.Chem., 62, 2019
|
|
6Q54
 
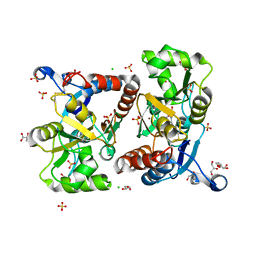 | | Structure of GluA2 ligand-binding domain (S1S2J) in complex with the agonist (S)-2-Amino-3-(1-ethyl-4-hydroxy-1H-1,2,3-triazol-5-yl)propanoic acid at 1.4 A resolution | | 分子名称: | (2~{S})-2-azanyl-3-(3-ethyl-5-oxidanyl-1,2,3-triazol-4-yl)propanoic acid, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Moellerud, S, Temperini, P, Kastrup, J.S. | | 登録日 | 2018-12-07 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Use of the 4-Hydroxytriazole Moiety as a Bioisosteric Tool in the Development of Ionotropic Glutamate Receptor Ligands.
J.Med.Chem., 62, 2019
|
|
6SBT
 
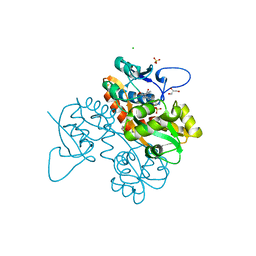 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with N-(7-(1H-imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl benzamide at 2.3 A resolution | | 分子名称: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | 著者 | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2019-07-22 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | N-(7-(1H-Imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)benzamide, a New Kainate Receptor Selective Antagonist and Analgesic: Synthesis, X-ray Crystallography, Structure-Affinity Relationships, and in Vitro and in Vivo Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
5NEB
 
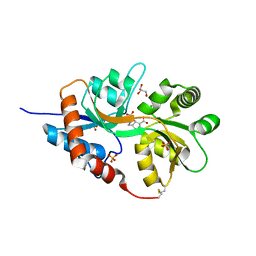 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with LM-12b at 2.05 A resolution | | 分子名称: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Moellerud, S, Frydenvang, K, Laulumaa, S, Kastrup, J.S. | | 登録日 | 2017-03-10 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5O4F
 
 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with the agonist LM-12b at 2.10 A resolution | | 分子名称: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-05-29 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
