7RK1
 
 | |
7RK2
 
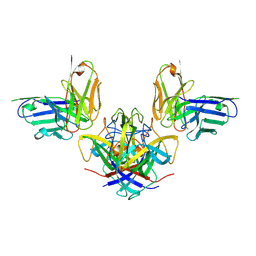 | | Crystal structure of the human astrovirus serotype 8 capsid spike in complex with scFv 2D9, an astrovirus-neutralizing antibody, at 2.65-A resolution | | 分子名称: | Capsid protein VP25, scFv 2D9 | | 著者 | Meyer, L, Cuellar, C, DuBois, R.M. | | 登録日 | 2021-07-21 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structures of Two Human Astrovirus Capsid/Neutralizing Antibody Complexes Reveal Distinct Epitopes and Inhibition of Virus Attachment to Cells.
J.Virol., 96, 2022
|
|
6HB3
 
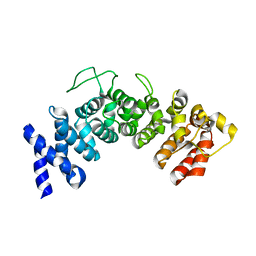 | | Structure of Hgh1, crystal form II | | 分子名称: | Protein HGH1 | | 著者 | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | 登録日 | 2018-08-09 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
6HB1
 
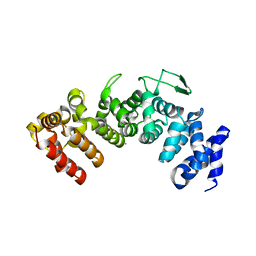 | | Structure of Hgh1, crystal form I | | 分子名称: | CHLORIDE ION, Protein HGH1 | | 著者 | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | 登録日 | 2018-08-09 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
6HB2
 
 | | Structure of Hgh1, crystal form I, Selenomethionine derivative | | 分子名称: | CHLORIDE ION, Protein HGH1 | | 著者 | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | 登録日 | 2018-08-09 | | 公開日 | 2019-02-27 | | 最終更新日 | 2019-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
1SR3
 
 | |
1KNG
 
 | | Crystal structure of CcmG reducing oxidoreductase at 1.14 A | | 分子名称: | THIOL:DISULFIDE INTERCHANGE PROTEIN CYCY | | 著者 | Edeling, M.A, Guddat, L.W, Fabianek, R.A, Thony-Meyer, L, Martin, J.L. | | 登録日 | 2001-12-18 | | 公開日 | 2002-07-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment
Structure, 10, 2002
|
|
1DT1
 
 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | 分子名称: | CYTOCHROME C552, HEME C | | 著者 | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | 登録日 | 2000-01-10 | | 公開日 | 2000-02-18 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
3BCI
 
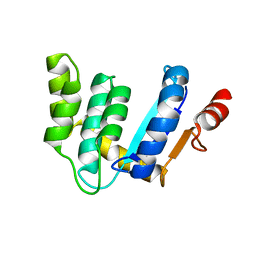 | |
3BCK
 
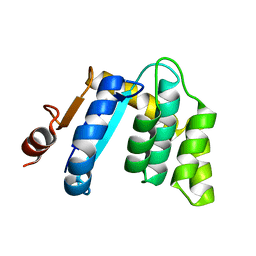 | |
3BD2
 
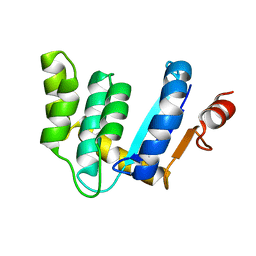 | |
7ZU0
 
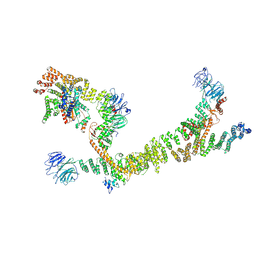 | | HOPS tethering complex from yeast | | 分子名称: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar morphogenesis protein 6, ... | | 著者 | Shvarev, D, Schoppe, J, Koenig, C, Perz, A, Fuellbrunn, N, Kiontke, S, Langemeyer, L, Januliene, D, Schnelle, K, Kuemmel, D, Froehlich, F, Moeller, A, Ungermann, C. | | 登録日 | 2022-05-11 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
7Z6K
 
 | |
7Z5Z
 
 | |
7Z6A
 
 | |
7Z5Y
 
 | |
1C7M
 
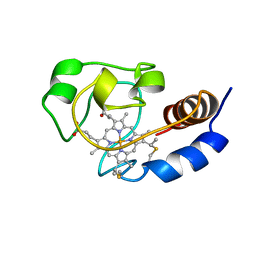 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | 分子名称: | HEME C, PROTEIN (CYTOCHROME C552) | | 著者 | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | 登録日 | 2000-03-03 | | 公開日 | 2000-07-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
2HL7
 
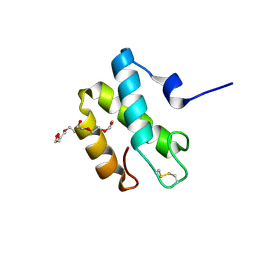 | | Crystal structure of the periplasmic domain of CcmH from Pseudomonas aeruginosa | | 分子名称: | Cytochrome C-type biogenesis protein CcmH, TETRAETHYLENE GLYCOL | | 著者 | Di Matteo, A, Travaglini-Allocatelli, C, Gianni, S, Brunori, M. | | 登録日 | 2006-07-06 | | 公開日 | 2007-07-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c
J.Biol.Chem., 282, 2007
|
|
4G9F
 
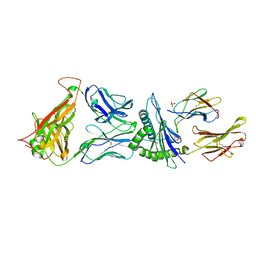 | | Crystal Structure of C12C TCR-HLAB2705-KK10-L6M | | 分子名称: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen, ... | | 著者 | Gras, S, Wilmann, P.G, Rossjohn, J. | | 登録日 | 2012-07-23 | | 公開日 | 2013-03-20 | | 最終更新日 | 2020-04-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4G8G
 
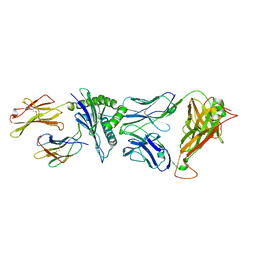 | | Crystal Structure of C12C TCR-HA B2705-KK10 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | 著者 | Gras, S, Wilmann, P.G, Rossjohn, J. | | 登録日 | 2012-07-23 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4G9D
 
 | | Crystal Structure of HLA B2705-KK10 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | 著者 | Gras, S, Wilmann, P.G, Rossjohn, J. | | 登録日 | 2012-07-23 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
4G8I
 
 | | Crystal Structure of HLA B2705-KK10-L6M | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, Gag protein, ... | | 著者 | Gras, S, Wilmann, P.G, Rossjohn, J. | | 登録日 | 2012-07-23 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
1I6E
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | 分子名称: | CYTOCHROME C552, HEME C | | 著者 | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | 登録日 | 2001-03-02 | | 公開日 | 2001-10-17 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1I6D
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | 分子名称: | CYTOCHROME C552, HEME C | | 著者 | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | 登録日 | 2001-03-02 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
7ZTY
 
 | |
