1KO7
 
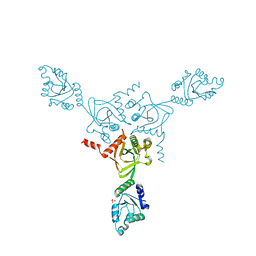 | | X-ray structure of the HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution | | 分子名称: | Hpr kinase/phosphatase, PHOSPHATE ION | | 著者 | Marquez, J.A, Hasenbein, S, Koch, B, Fieulaine, S, Nessler, S, Hengstenberg, W, Scheffzek, K. | | 登録日 | 2001-12-20 | | 公開日 | 2002-04-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2NMS
 
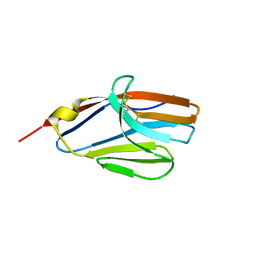 | |
5JXP
 
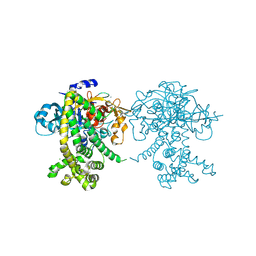 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | 分子名称: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | 著者 | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | 登録日 | 2016-05-13 | | 公開日 | 2017-06-14 | | 最終更新日 | 2017-06-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
4QGK
 
 | |
3QN1
 
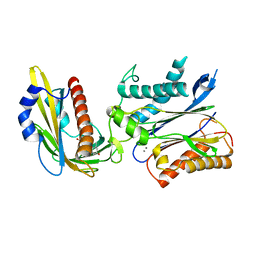 | | Crystal structure of the PYR1 Abscisic Acid receptor in complex with the HAB1 type 2C phosphatase catalytic domain | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | 著者 | Betz, K, Dupeux, F, Santiago, J, Marquez, J.A. | | 登録日 | 2011-02-07 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele.
Plant Physiol., 156, 2011
|
|
3K90
 
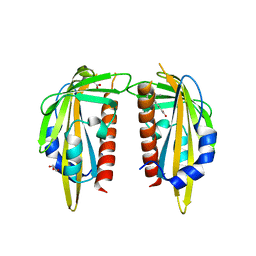 | | The Abscisic acid receptor PYR1 in complex with Abscisic Acid | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ACETIC ACID, GLYCEROL, ... | | 著者 | Dupeux, F.D, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | 登録日 | 2009-10-15 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The abscisic acid receptor PYR1 in complex with abscisic acid.
Nature, 462, 2009
|
|
2Q87
 
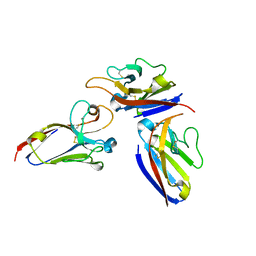 | |
3ZVU
 
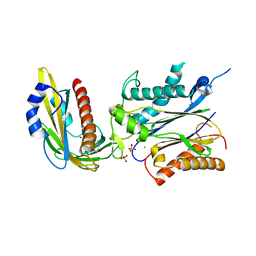 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | 著者 | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | 登録日 | 2011-07-27 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
4AXT
 
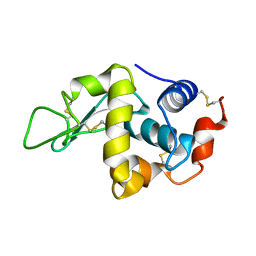 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME FROM AN AUTO HARVESTED CRYSTAL, Control Experiment | | 分子名称: | LYSOZYME C | | 著者 | Cipriani, F, Rower, M, Landret, C, Zander, U, Felisaz, F, Marquez, J.A. | | 登録日 | 2012-06-14 | | 公開日 | 2012-10-24 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Crystal Direct: A New Method for Automated Crystal Harvesting Based on Laser-Induced Photoablation of Thin Films
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5DWP
 
 | | Crystal Structure of human transthyretin (TTR) processed with the CrystalDirect automated mounting and cryo-cooling technology | | 分子名称: | Transthyretin | | 著者 | Zander, U, Cornaciu, I, Forsyth, T, Yee, A, Marquez, J.A. | | 登録日 | 2015-09-22 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Automated harvesting and processing of protein crystals through laser photoablation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EBH
 
 | |
1RJ1
 
 | | Crystal Structure of a Cell Wall Invertase Inhibitor from Tobacco | | 分子名称: | invertase inhibitor | | 著者 | Hothorn, M, D'Angelo, I, Marquez, J.A, Greiner, S, Scheffzek, K. | | 登録日 | 2003-11-18 | | 公開日 | 2004-02-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension
J.Mol.Biol., 335, 2004
|
|
1RJ4
 
 | | Structure of a Cell Wall Invertase Inhibitor from Tobacco in Complex with Cd2+ | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, invertase inhibitor | | 著者 | Hothorn, M, D'Angelo, I, Marquez, J.A, Greiner, S, Scheffzek, K. | | 登録日 | 2003-11-18 | | 公開日 | 2004-02-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension
J.Mol.Biol., 335, 2004
|
|
8QQ1
 
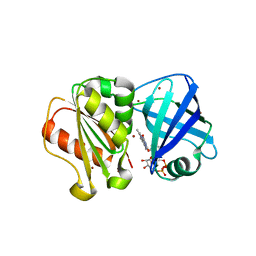 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | 分子名称: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | 著者 | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | 登録日 | 2023-10-03 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
6FZS
 
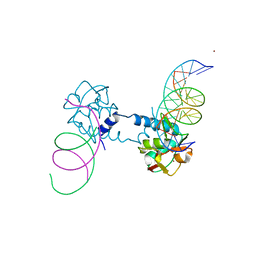 | |
6FZT
 
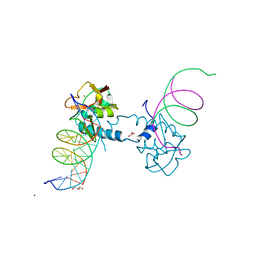 | | Crystal structure of Smad8_9-MH1 bound to the GGCGC site. | | 分子名称: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), GLYCEROL, Mothers against decapentaplegic homolog 9, ... | | 著者 | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | 登録日 | 2018-03-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
6H3R
 
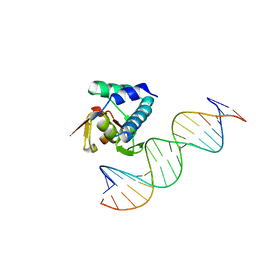 | |
2X89
 
 | | Structure of the Beta2_microglobulin involved in amyloidogenesis | | 分子名称: | ANTIBODY, BETA-2-MICROGLOBULIN | | 著者 | Domanska, K, Srinivasan, V, Vanderhaegen, S, Pardon, E, Marquez, J.A, Bellotti, V, Wyns, L, Steyaert, J. | | 登録日 | 2010-03-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Atomic Structure of a Nanobody-Trapped Domain-Swapped Dimer of an Amyloidogenic {Beta}2-Microglobulin Variant.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4C9C
 
 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 1E protein (Form A) | | 分子名称: | GLYCEROL, MAJOR STRAWBERRY ALLERGEN FRA A 1-E, SULFATE ION | | 著者 | Casanal, A, Zander, U, Valpuesta, V, Marquez, J.A. | | 登録日 | 2013-10-02 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Strawberry Pathogenesis-Related 10 (Pr-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates.
J.Biol.Chem., 288, 2013
|
|
4C94
 
 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 3 protein in complex with Catechin | | 分子名称: | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, FRA A 3 ALLERGEN | | 著者 | Casanal, A, Zander, U, Valpuesta, V, Marquez, J.A. | | 登録日 | 2013-10-02 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Strawberry Pathogenesis-Related 10 (Pr-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates.
J.Biol.Chem., 288, 2013
|
|
5LKU
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with coenzyme A. | | 分子名称: | COENZYME A, Histone acetyltransferase p300,Histone acetyltransferase p300, ZINC ION | | 著者 | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | 登録日 | 2016-07-25 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LKT
 
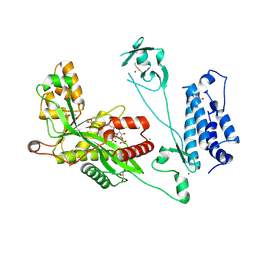 | | Crystal structure of the p300 acetyltransferase catalytic core with butyryl-coenzyme A. | | 分子名称: | Butyryl Coenzyme A, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | 登録日 | 2016-07-24 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LKZ
 
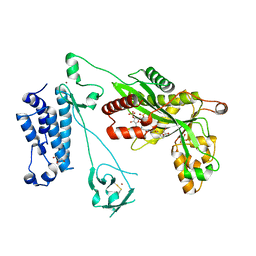 | | Crystal structure of the p300 acetyltransferase catalytic core with crotonyl-coenzyme A. | | 分子名称: | CROTONYL COENZYME A, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | 著者 | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | 登録日 | 2016-07-25 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5M7N
 
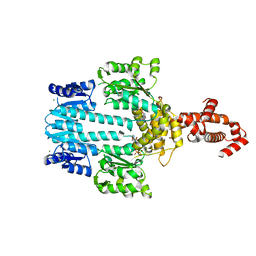 | | Crystal structure of NtrX from Brucella abortus in complex with ATP processed with the CrystalDirect automated mounting and cryo-cooling technology | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | 著者 | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | 登録日 | 2016-10-28 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
5M7P
 
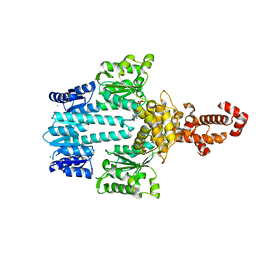 | | Crystal structure of NtrX from Brucella abortus in complex with ADP processed with the CrystalDirect automated mounting and cryo-cooling technology | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | 著者 | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | 登録日 | 2016-10-28 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
