3HNI
 
 | | Crystal structure of the Zn-induced tetramer of the engineered cyt cb562 variant RIDC-1 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ZINC ION | | 著者 | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | 登録日 | 2009-05-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HNK
 
 | | Crystal structure of the dimeric assembly of the cyt cb562 variant RIDC-1 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | 著者 | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | 登録日 | 2009-05-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HNJ
 
 | | Crystal structure of the Zn-induced tetramer of the engineered cyt cb562 variant RIDC-2 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ZINC ION | | 著者 | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | 登録日 | 2009-05-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HNL
 
 | | Crystal structure of the Cu-induced dimer of the engineered cyt cb562 variant RIDC-1 | | 分子名称: | CHLORIDE ION, COPPER (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Salgado, E.N, Lewis, R.A, Brodin, J, Tezcan, F.A. | | 登録日 | 2009-05-31 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Metal templated design of protein interfaces.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3C63
 
 | | Tetrameric Cytochrome cb562 (K34/H59/D62/H63/H73/A74/H77) Assembly Stabilized by Interprotein Zinc Coordination | | 分子名称: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | 著者 | Tezcan, F.A, Salgado, E.N, Lewis, R.A, Faraone-Mennella, J. | | 登録日 | 2008-02-02 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
J.Am.Chem.Soc., 130, 2008
|
|
3C62
 
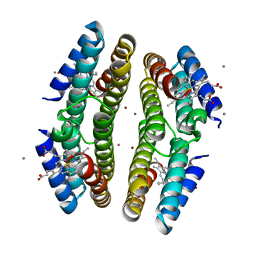 | | Tetrameric Cytochrome cb562 (H59/D62/H63/H73/A74/H77) Assembly Stabilized by Interprotein Zinc Coordination | | 分子名称: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | 著者 | Tezcan, F.A, Salgado, E.N, Lewis, R.A, Faraone-Mennella, J. | | 登録日 | 2008-02-02 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
J.Am.Chem.Soc., 130, 2008
|
|
3DE8
 
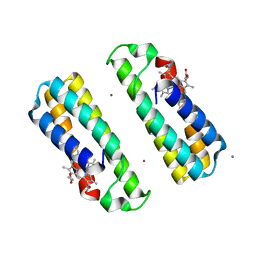 | | Crystal Structure of a Dimeric Cytochrome cb562 Assembly Induced by Copper Coordination | | 分子名称: | CALCIUM ION, COPPER (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Salgado, E.N, Lewis, R.A, Rheingold, A.L, Tezcan, F.A. | | 登録日 | 2008-06-08 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through Cu(II) and Ni(II) coordination.
Inorg.Chem., 48, 2009
|
|
3DE9
 
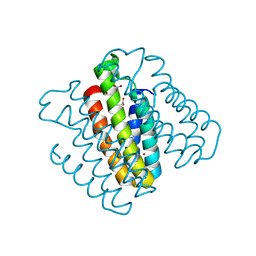 | | Crystal Structure of a Trimeric Cytochrome cb562 Assembly Induced by Nickel Coordination | | 分子名称: | NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | 著者 | Salgado, E.N, Lewis, R.A, Rheingold, A.L, Tezcan, F.A. | | 登録日 | 2008-06-08 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through Cu(II) and Ni(II) coordination.
Inorg.Chem., 48, 2009
|
|
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | 分子名称: | Uncharacterized protein | | 著者 | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | 登録日 | 2013-04-06 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
2M6P
 
 | |
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | 分子名称: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDC
 
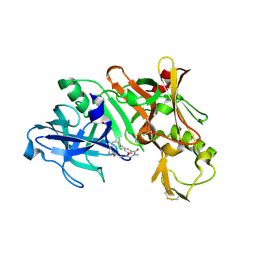 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | 分子名称: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD2
 
 | | Crystal structure of BACE complex with BMC017 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCW
 
 | | Crystal structure of BACE complex with BMC021 | | 分子名称: | (2R,4S)-N-butyl-4-[(2S,5S,7R)-2,7-dimethyl-3,15-dioxo-1,4-diazacyclopentadecan-5-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDA
 
 | | Crystal structure of BACE complex with BMC013 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCO
 
 | | Crystal structure of BACE complex with BMC016 | | 分子名称: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD7
 
 | | Crystal structure of BACE complex with BMC014 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCQ
 
 | | Crystal structure of BACE complex with BMC025 | | 分子名称: | (2R,4S,5S)-N-butyl-4-hydroxy-2,7-dimethyl-5-{[N-(4-methylpentanoyl)-L-methionyl]amino}octanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDD
 
 | | Crystal structure of BACE complex with BMC020 hydrolyzed | | 分子名称: | (10R,12S)-12-[(1R)-1,2-dihydroxyethyl]-N,N,10-trimethyl-14-oxo-2-oxa-13-azabicyclo[13.3.1]nonadeca-1(19),15,17-triene-17-carboxamide, Beta-secretase 1, GLYCEROL | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD3
 
 | | Crystal structure of BACE complex with BMC010 | | 分子名称: | (10R,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCY
 
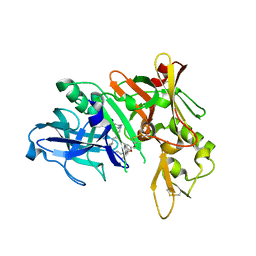 | | Crystal structure of BACE complex with BMC008 | | 分子名称: | (9R,11S)-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-16-(1,3-oxazol-2-yl)-3-[(1R)-1-phenylethyl]-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCX
 
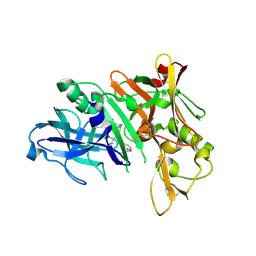 | | Crystal structure of BACE complex with BMC007 | | 分子名称: | (9R,11S)-3-ethyl-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD9
 
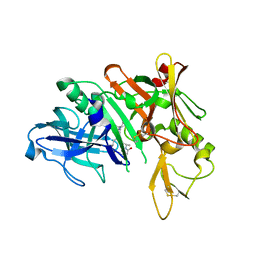 | | Crystal structure of BACE complex with BMC005 | | 分子名称: | (5S,8S,10R)-8-[(1R)-2-{[1-(3-tert-butylphenyl)cyclopropyl]amino}-1-hydroxyethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCP
 
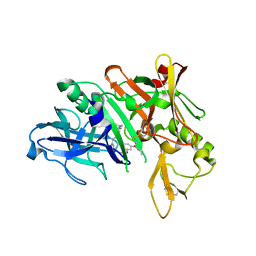 | | Crystal structure of BACE complex with BMC018 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(2-oxopropoxy)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD5
 
 | | Crystal structure of BACE complex with BMC009 | | 分子名称: | (10S,12S)-17-chloro-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-10-methyl-7-oxa-2,13,18-triazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
