8SH5
 
 | | Crystal structure of 3'cap-independent translation enhancers (CITE) from Pea enation mosaic virus RNA 2 (PEMV2) with Fab BL3-6K170A | | 分子名称: | Fab BL3-6K170A heavy chain, Fab BL3-6K170A light chain, RNA (88-MER) | | 著者 | Lewicka, A, Roman, C, Rice, P.A, Piccirilli, J.A. | | 登録日 | 2023-04-13 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of a cap-independent translation enhancer RNA.
Nucleic Acids Res., 51, 2023
|
|
7MLX
 
 | |
8UTA
 
 | | yjdF riboswitch from R. gauvreauii in complex with proflavine bound to Fab BL3-6 S97N | | 分子名称: | Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, MAGNESIUM ION, ... | | 著者 | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
8UIW
 
 | | yjdF riboswitch from R. gauvreauii in complex with chelerythrine bound to Fab BL3-6 S97N | | 分子名称: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, ... | | 著者 | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | 登録日 | 2023-10-10 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | 分子名称: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | 著者 | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | 登録日 | 2019-09-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6U8D
 
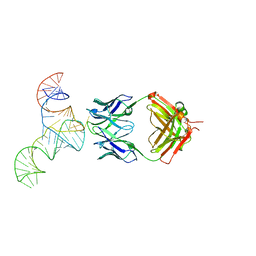 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | 分子名称: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | 著者 | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | 登録日 | 2019-09-04 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.807 Å) | | 主引用文献 | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
