1EFS
 
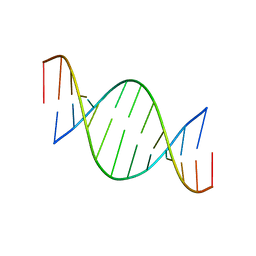 | | CONFORMATION OF A DNA-RNA HYBRID | | 分子名称: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*UP*CP*UP*UP*CP*CP*UP*CP*UP*C)-3') | | 著者 | Larue, V, Hantz, E. | | 登録日 | 2000-02-10 | | 公開日 | 2000-03-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution conformation of an RNA--DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand.
Int.J.Biol.Macromol., 28, 2001
|
|
6Q56
 
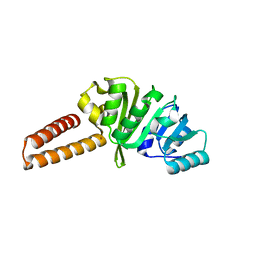 | | Crystal structure of the B. subtilis M1A22 tRNA methyltransferase TrmK | | 分子名称: | NICKEL (II) ION, tRNA (adenine(22)-N(1))-methyltransferase | | 著者 | Degut, C, Roovers, M, Barraud, P, Brachet, F, Feller, A, Larue, V, Al Refaii, A, Caillet, J, Droogmans, L, Tisne, C. | | 登録日 | 2018-12-07 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural characterization of B. subtilis m1A22 tRNA methyltransferase TrmK: insights into tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
6RWG
 
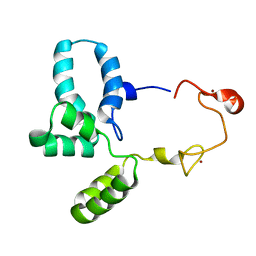 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | 分子名称: | Gag polyprotein, ZINC ION | | 著者 | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-05 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
4JZS
 
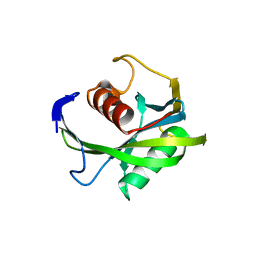 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZU
 
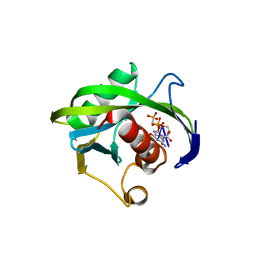 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZT
 
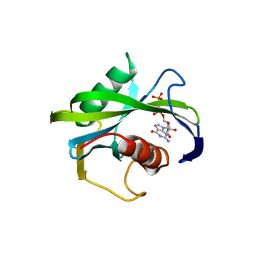 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZV
 
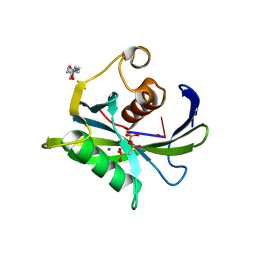 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GSH
 
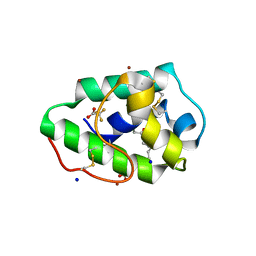 | | Three-dimensional structure of a post translational modified barley LTP1 | | 分子名称: | (12E)-10-oxooctadec-12-enoic acid, Non-specific lipid-transfer protein 1, SODIUM ION, ... | | 著者 | Lascombe, M.B, Prange, T, Bakan, B, Marion, D. | | 登録日 | 2009-03-27 | | 公開日 | 2009-12-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of oxylipin-conjugated barley LTP1 highlights the unique plasticity of the hydrophobic cavity of these plant lipid-binding proteins.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
2RKN
 
 | | X-ray structure of the self-defense and signaling protein DIR1 from Arabidopsis taliana | | 分子名称: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, DIR1 protein, ZINC ION | | 著者 | Lascombe, M.B, Prange, T, Buhot, N, Marion, D, Bakan, B, Lamb, C. | | 登録日 | 2007-10-17 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structure of "defective in induced resistance" protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein.
Protein Sci., 17, 2008
|
|
