4TX1
 
 | |
2L4M
 
 | |
2FI7
 
 | |
2HK1
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the presence of D-fructose | | 分子名称: | D-PSICOSE 3-EPIMERASE, D-fructose, MANGANESE (II) ION | | 著者 | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | 登録日 | 2006-07-03 | | 公開日 | 2006-08-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
2HK0
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the absence of substrate | | 分子名称: | D-PSICOSE 3-EPIMERASE | | 著者 | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | 登録日 | 2006-07-03 | | 公開日 | 2006-08-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
3E74
 
 | |
7N4Y
 
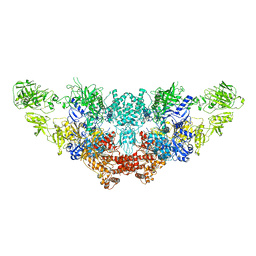 | | The structure of bovine thyroglobulin with iodinated tyrosines | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kim, K, Clarke, O.B. | | 登録日 | 2021-06-04 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | The structure of natively iodinated bovine thyroglobulin.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2Q37
 
 | |
7CMX
 
 | |
7CMY
 
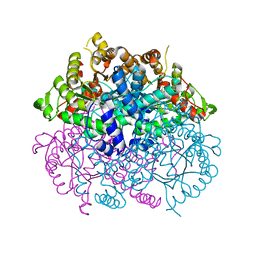 | | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate | | 分子名称: | GLYOXYLIC ACID, Isocitrate lyase, MAGNESIUM ION, ... | | 著者 | Kim, K, Ki, D, Lee, S.H. | | 登録日 | 2020-07-29 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate
To Be Published
|
|
1V9H
 
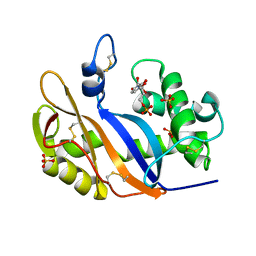 | | Crystal structure of the RNase MC1 mutant Y101A in complex with 5'-UMP | | 分子名称: | Ribonuclease MC, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | 著者 | Kimura, K, Numata, T, Kakuta, Y, Kimura, M. | | 登録日 | 2004-01-26 | | 公開日 | 2004-10-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Amino acids conserved at the C-terminal half of the ribonuclease t2 family contribute to protein stability of the enzymes
Biosci.Biotechnol.Biochem., 68, 2004
|
|
1VE8
 
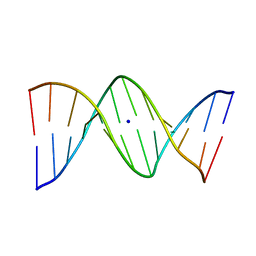 | | X-Ray analyses of oligonucleotides containing 5-formylcytosine, suggesting a structural reason for codon-anticodon recognition of mitochondrial tRNA-Met; Part 1, d(CGCGAATT(f5C)GCG) | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5FC)P*GP*CP*G)-3', SODIUM ION | | 著者 | Kimura, K, Ono, A, Watanabe, K, Takenaka, A. | | 登録日 | 2004-03-29 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | X-Ray analyses of oligonucleotides containing 5-formylcytosine, suggest a structural reason for the codon-anticodon recognition of mitochondrial tRNA-Met
To be Published
|
|
7ECD
 
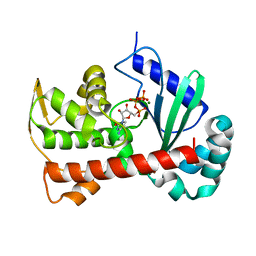 | | Crystal structure of Tam41 from Firmicutes bacterium, complex with CTP-Mg | | 分子名称: | BROMIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Kimura, K, Kawai, F, Kubota-Kawai, H, Watanabe, Y, Tamura, Y. | | 登録日 | 2021-03-12 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Tam41 cytidine diphosphate diacylglycerol synthase from a Firmicutes bacterium.
J.Biochem., 171, 2022
|
|
3ACF
 
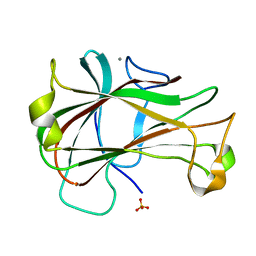 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACG
 
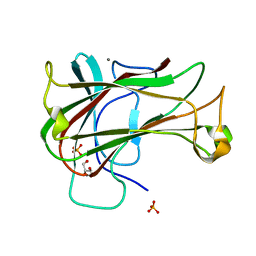 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellobiose | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, GLYCEROL, ... | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACI
 
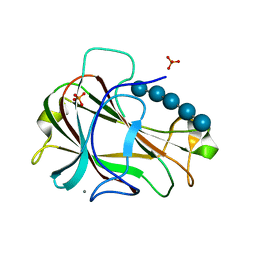 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellopentaose | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
3ACH
 
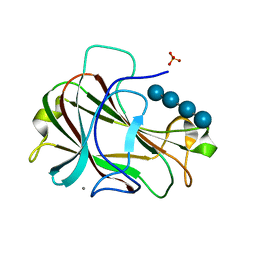 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in complex with cellotetraose | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
6WHA
 
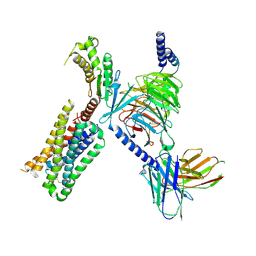 | | HTR2A bound to 25-CN-NBOH in complex with a mini-Galpha-q protein, beta/gamma subunits and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | 分子名称: | 4-(2-{[(2-hydroxyphenyl)methyl]amino}ethyl)-2,5-dimethoxybenzonitrile, G subunit q (Gi2-mini-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Skiniotis, G, Roth, B, Kim, K, Panova, O. | | 登録日 | 2020-04-07 | | 公開日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
7YIT
 
 | |
5W7W
 
 | |
5W7Y
 
 | |
5W7X
 
 | |
3WNP
 
 | | D308A, F268V, D469Y, A513V, and Y515S quintuple mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoundecaose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | 著者 | Suzuki, R, Suzuki, N, Fujimoto, Z, Momma, M, Kimura, K, Kitamura, S, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular engineering of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040: structural determinants for the reaction product size and reactivity.
Biochem.J., 467, 2015
|
|
7SRR
 
 | | 5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM) | | 分子名称: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2B, G protein subunit q (Gi2-mini-Gq chimera), ... | | 著者 | Barros-Alvarez, X, Kim, K, Panova, O, Cao, C, Roth, B.L, Skiniotis, G. | | 登録日 | 2021-11-08 | | 公開日 | 2022-09-21 | | 最終更新日 | 2022-10-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
4R3P
 
 | | Crystal structures of EGFR in complex with Mig6 | | 分子名称: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | 著者 | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | 登録日 | 2014-08-17 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.905 Å) | | 主引用文献 | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
