8U5F
 
 | | Crystal Structure of Trypsinized Clostridium perfringens Enterotoxin | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Heat-labile enterotoxin B chain, ... | | 著者 | Kapoor, S, Ogbu, C.P, Vecchio, A.J. | | 登録日 | 2023-09-12 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin.
Toxins, 15, 2023
|
|
7CT3
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | 分子名称: | Mutual gliding motility protein C (MglC), SODIUM ION | | 著者 | Thakur, K.G, Kapoor, S, Kodesia, A. | | 登録日 | 2020-08-17 | | 公開日 | 2021-01-27 | | 最終更新日 | 2021-07-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 296, 2021
|
|
7CY1
 
 | |
8U5E
 
 | |
8U5D
 
 | |
7XQC
 
 | |
4QGS
 
 | |
5I2A
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diol-dehydratase | | 著者 | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | 登録日 | 2016-02-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
5I2G
 
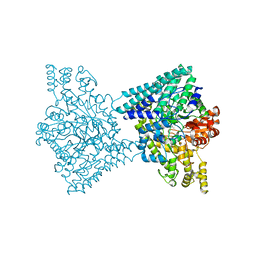 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | 分子名称: | Diol dehydratase, S-1,2-PROPANEDIOL | | 著者 | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | 登録日 | 2016-02-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.352 Å) | | 主引用文献 | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
5ZFQ
 
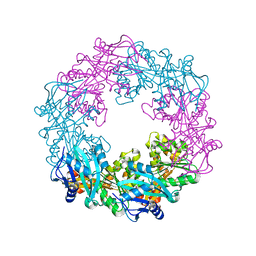 | |
5ZFR
 
 | | Crystal structure of PilB, an extension ATPase motor of Type IV pilus, from Geobacter sulfurreducens | | 分子名称: | PHOSPHATE ION, Type IV pilus biogenesis ATPase PilB, ZINC ION | | 著者 | Thakur, K.G, Solanki, V, Kapoor, S. | | 登録日 | 2018-03-06 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural insights into the mechanism of Type IVa pilus extension and retraction ATPase motors
FEBS J., 285, 2018
|
|
8OYS
 
 | |
8OYV
 
 | |
8OYX
 
 | |
8OYW
 
 | |
8OYY
 
 | |
9BEI
 
 | |
2VHY
 
 | |
2VHX
 
 | |
2VHZ
 
 | |
2VHW
 
 | |
2VHV
 
 | |
