6PLN
 
 | |
6XJF
 
 | |
5FJG
 
 | | The crystal structure of light-driven chloride pump ClR in pH 4.5. | | 分子名称: | ANHYDRORETINOL, CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2015-10-07 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structure of Light-Driven Chloride Pump Clr in Ph 4.5.
To be Published
|
|
6KF3
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF9
 
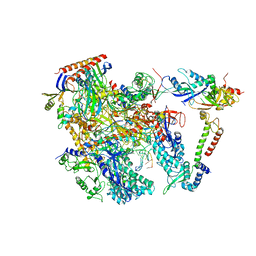 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-07 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
4QIW
 
 | |
4QJV
 
 | |
7VWX
 
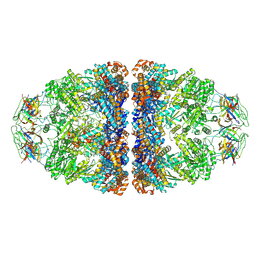 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | 分子名称: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | 著者 | Kim, H, Roh, S.H. | | 登録日 | 2021-11-12 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
2OLE
 
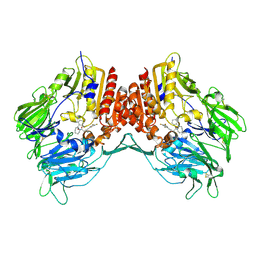 | | Crystal Structure Of Human Dipeptidyl Peptidase IV (DPPIV) Complex With Cyclic Hydrazine Derivatives | | 分子名称: | (2R)-4-(2-BENZOYL-1,2-DIAZEPAN-1-YL)-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE, Dipeptidyl peptidase 4 | | 著者 | Kim, S.S, Ahn, J.H, Lee, J.O. | | 登録日 | 2007-01-19 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Synthesis, biological evaluation and structural determination of beta-aminoacyl-containing cyclic hydrazine derivatives as dipeptidyl peptidase IV (DPP-IV) inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8GW7
 
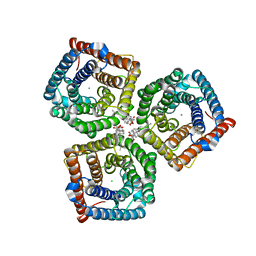 | | AtSLAC1 6D mutant in open state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2022-09-16 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW6
 
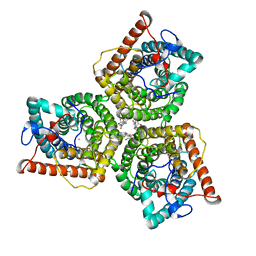 | | AtSLAC1 6D mutant in closed state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2022-09-16 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
5G2A
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0 with Bromide ion. | | 分子名称: | BROMIDE ION, CHLORIDE PUMPING RHODOPSIN, RETINAL | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G54
 
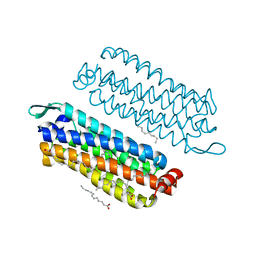 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-05-19 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | 分子名称: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | 分子名称: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | 分子名称: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5H62
 
 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H60
 
 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | MANGANESE (II) ION, Transferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-20 | | 最終更新日 | 2018-10-31 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H61
 
 | |
5H5Y
 
 | |
5H63
 
 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | MANGANESE (II) ION, Transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2023-04-11 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J1E
 
 | | AtSLAC1 in open state | | 分子名称: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | 著者 | Lee, Y, Lee, S. | | 登録日 | 2023-04-12 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
