5VXE
 
 | | Crystal structure of Xanthomonas campestris OleA E117A bound with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Wilmot, C.M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
5VXD
 
 | |
5VXI
 
 | | Crystal structure of Xanthomonas campestris OleA E117D bound with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Wilmot, C.M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
5VXF
 
 | |
5VXH
 
 | |
5VXG
 
 | | Crystal structure of Xanthomonas campestris OleA E117Q bound with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Wilmot, C.M. | | 登録日 | 2017-05-23 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | OleA Glu117 is key to condensation of two fatty-acyl coenzyme A substrates in long-chain olefin biosynthesis.
Biochem. J., 474, 2017
|
|
6B2S
 
 | | Crystal structure of Xanthomonas campestris OleA H285N | | 分子名称: | 3-oxoacyl-[ACP] synthase III, GLYCEROL, PHOSPHATE ION | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2R
 
 | | Crystal structure of Xanthomonas campestris OleA H285A | | 分子名称: | 3-oxoacyl-[ACP] synthase III, GLYCEROL | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2T
 
 | | Crystal structure of Xanthomonas campestris OleA H285D | | 分子名称: | 3-oxoacyl-[ACP] synthase III, GLYCEROL, PHOSPHATE ION | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2U
 
 | | Crystal structure of Xanthomonas campestris OleA H285N with Cerulenin | | 分子名称: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | 著者 | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | 登録日 | 2017-09-20 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The role of OleA His285 in substrate coordination of long-chain acyl-CoA
To be published
|
|
3SXT
 
 | |
3SWS
 
 | |
3RN1
 
 | |
4XW2
 
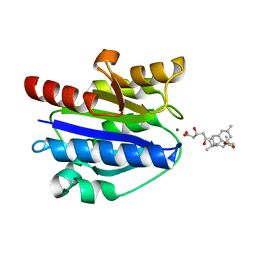 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | 分子名称: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | 著者 | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | 登録日 | 2015-01-28 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
7NYO
 
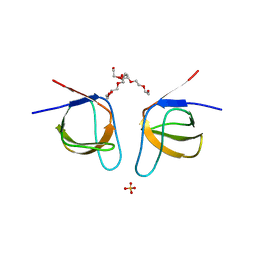 | | Mutant A541L of SH3 domain of JNK-interacting Protein 1 (JIP1) | | 分子名称: | 1,2-ETHANEDIOL, SH3 domain of JNK-interacting Protein 1 (JIP1), SULFATE ION, ... | | 著者 | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | 登録日 | 2021-03-23 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZC
 
 | |
7NYK
 
 | |
7NYM
 
 | | Mutant V517A - SH3 domain of JNK-interacting Protein 1 (JIP1) | | 分子名称: | HEXAETHYLENE GLYCOL, PHOSPHATE ION, SH3 domain of JNK-interacting Protein 1 (JIP1), ... | | 著者 | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | 登録日 | 2021-03-23 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.614 Å) | | 主引用文献 | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZB
 
 | | Mutant V517L of the SH3 domain of JNK-interacting protein 1 (JIP1) | | 分子名称: | PHOSPHATE ION, SH3 domain of JNK-interacting protein 1 (JIP1), TETRAETHYLENE GLYCOL | | 著者 | Perez, L.M, Ielasi, F.S, Jensen, M.R, Palencia, A. | | 登録日 | 2021-03-23 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.959 Å) | | 主引用文献 | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | 著者 | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | 登録日 | 2021-03-23 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.537 Å) | | 主引用文献 | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYL
 
 | | Mutant H493A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | 分子名称: | SH3 domain of JNK-interacting Protein 1 (JIP1), TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | 著者 | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | 登録日 | 2021-03-23 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZD
 
 | | Fourth SH3 domain of POSH (Plenty of SH3 Domains protein) | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase SH3RF1 | | 著者 | Palencia, A, Bessa, L.M, Jensen, M.R. | | 登録日 | 2021-03-23 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
8RPP
 
 | |
2K6U
 
 | | The Solution Structure of a Conformationally Restricted Fully Active Derivative of the Human Relaxin-like Factor (RLF) | | 分子名称: | Insulin-like 3 A chain, Insulin-like 3 B chain | | 著者 | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | 登録日 | 2008-07-24 | | 公開日 | 2008-12-16 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
2K6T
 
 | | Solution structure of the relaxin-like factor | | 分子名称: | Insulin-like 3 A chain, Insulin-like 3 B chain | | 著者 | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | 登録日 | 2008-07-23 | | 公開日 | 2008-12-16 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
