3AH7
 
 | | Crystal structure of the ISC-like [2Fe-2S] ferredoxin (FdxB) from Pseudomonas putida JCM 20004 | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, SODIUM ION, ... | | 著者 | Kumasaka, T, Shimizu, N, Ohmori, D, Iwasaki, T. | | 登録日 | 2010-04-20 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the ISC-like [2Fe-2S] ferredoxin (FdxB) from Pseudomonas putida JCM 20004
To be Published
|
|
5YDN
 
 | | Mu pahge neck subunit | | 分子名称: | Gene product J | | 著者 | Takeda, S, Iwasaki, T, Yamashita, E, Nakagawa, A. | | 登録日 | 2017-09-13 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Three-dimensional structures of bacteriophage neck subunits are shared in Podoviridae, Siphoviridae and Myoviridae
Genes Cells, 23, 2018
|
|
7YVZ
 
 | | Structure of Caenorhabditis elegans CISD-1/mitoNEET | | 分子名称: | CDGSH iron-sulfur domain-containing protein 1 (CISD-1/mitoNEET), FE2/S2 (INORGANIC) CLUSTER | | 著者 | Hasegawa, K, Hagiuda, E, Taguchi, A.T, Geldenhuys, W, Iwasaki, T, Kumasaka, T. | | 登録日 | 2022-08-20 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and biological evaluation of Caenorhabditis elegans CISD-1/mitoNEET, a KLP-17 tail domain homologue, supports attenuation of paraquat-induced oxidative stress through a p38 MAPK-mediated antioxidant defense response.
Adv Redox Res, 6, 2022
|
|
6DE9
 
 | | mitoNEET bound to furosemide | | 分子名称: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | 著者 | Robart, A.R, Geldenhuys, W.J. | | 登録日 | 2018-05-11 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the mitochondrial protein mitoNEET bound to a benze-sulfonide ligand.
Commun Chem, 2, 2019
|
|
5GXQ
 
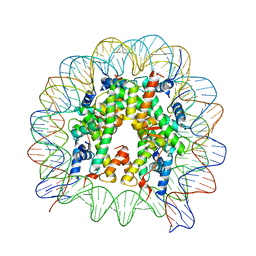 | | The crystal structure of the nucleosome containing H3.6 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2016-09-19 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
7C25
 
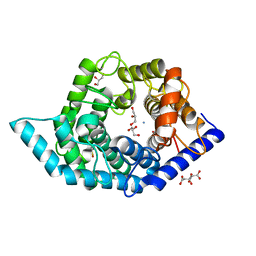 | | Glycosidase Wild Type at pH8.0 | | 分子名称: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.505 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C24
 
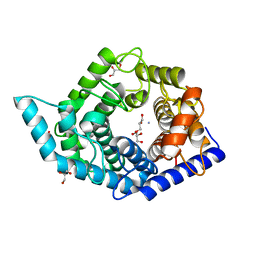 | | Glycosidase F290Y at pH8.0 | | 分子名称: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C26
 
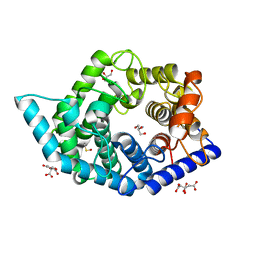 | | Glycosidase Wild Type at pH4.5 | | 分子名称: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C27
 
 | | Glycosidase F290Y at pH4.5 | | 分子名称: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
6JU1
 
 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | 分子名称: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | 著者 | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | 登録日 | 2019-04-12 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
5XM0
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.874 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5X7X
 
 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Arimura, Y, Taguchi, H, Kurumizaka, H. | | 登録日 | 2017-02-27 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.184 Å) | | 主引用文献 | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
5XM1
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5Z3D
 
 | | Glycosidase F290Y | | 分子名称: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2022-02-23 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3C
 
 | | Glycosidase E178A | | 分子名称: | GLYCEROL, Glycoside hydrolase 15-related protein | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3B
 
 | | Glycosidase Y48F | | 分子名称: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3A
 
 | | Glycosidase Wild Type | | 分子名称: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3F
 
 | | Glycosidase E335A in complex with glucose | | 分子名称: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein, ... | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
5Z3E
 
 | | Glycosidase E335A | | 分子名称: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
