4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | 分子名称: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | 著者 | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | 登録日 | 2014-11-17 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TKP
 
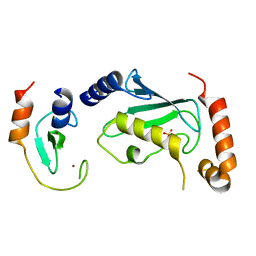 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | 分子名称: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | 著者 | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | 登録日 | 2014-05-27 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
2LM3
 
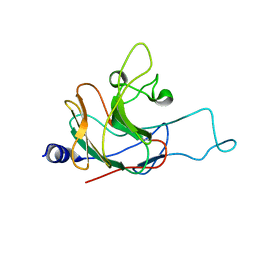 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | 分子名称: | Tripartite motif-containing protein 5 | | 著者 | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | 登録日 | 2011-11-21 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6U6Z
 
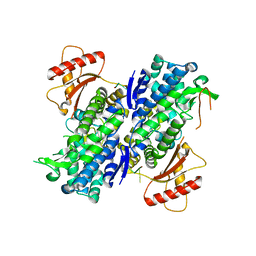 | |
6U6X
 
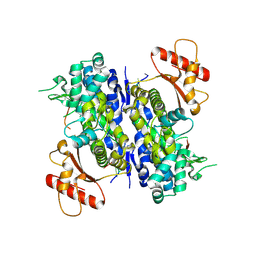 | | Human SAMHD1 bound to deoxyribo(C*G*C*C*T)-oligonucleotide | | 分子名称: | DNA SC-GS-SC-SC-DT, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | 著者 | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | 登録日 | 2019-08-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
6U6Y
 
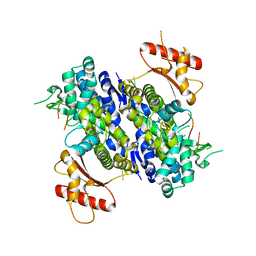 | | Human SAMHD1 bound to ribo(CGCCU)-oligonucleotide | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, RNA CGCCU, ... | | 著者 | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | 登録日 | 2019-08-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
3UV9
 
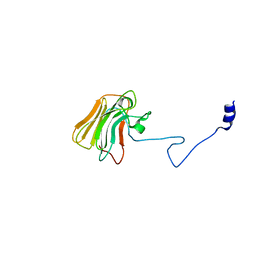 | | Structure of the rhesus monkey TRIM5alpha deltav1 PRYSPRY domain | | 分子名称: | Tripartite motif-containing protein 5 | | 著者 | Biris, N, Yang, Y, Taylor, A.B, Tomashevskii, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D. | | 登録日 | 2011-11-29 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JVW
 
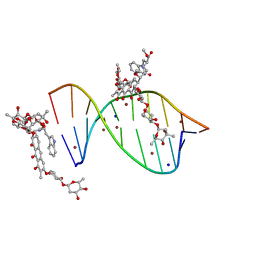 | | Crystal structure of mithramycin analogue MTM SA-Trp in complex with a 10-mer DNA AGAGGCCTCT. | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Trp, ... | | 著者 | Hou, C, Rohr, J, Tsodikov, O.V. | | 登録日 | 2016-05-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
5VA4
 
 | |
7JSA
 
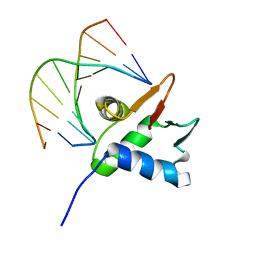 | |
7JSL
 
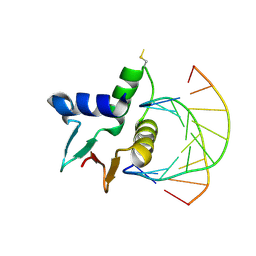 | |
7LU5
 
 | | SAMHD1(113-626) H206R D207N R366H | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | 著者 | Temple, J.T, Bowen, N.E. | | 登録日 | 2021-02-20 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.57 Å) | | 主引用文献 | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
7LTT
 
 | | SAMHD1(113-626) H206R D207N R366C | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | 著者 | Temple, J.T, Bowen, N.E. | | 登録日 | 2021-02-20 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
4QNB
 
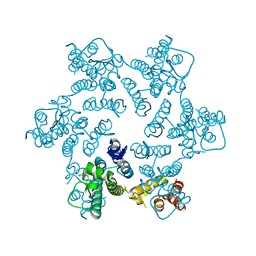 | |
6OW0
 
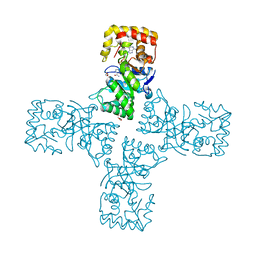 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+ and PEG | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MtmW, ... | | 著者 | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | 登録日 | 2019-05-08 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OVX
 
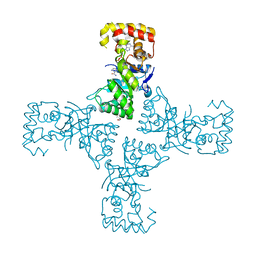 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | 分子名称: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | 著者 | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | 登録日 | 2019-05-08 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OVQ
 
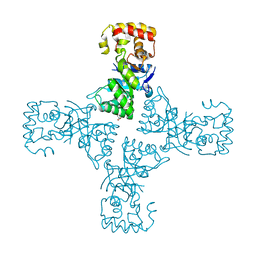 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | 分子名称: | GLYCEROL, Putative Side chain reductase | | 著者 | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | 登録日 | 2019-05-08 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5JW0
 
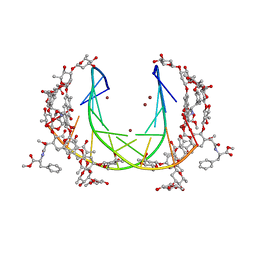 | | Crystal structure of mithramycin analogue MTM SA-Phe in complex with a 10-mer DNA AGGGTACCCT | | 分子名称: | DNA (5'-D(P*AP*GP*GP*GP*TP*AP*CP*CP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Phe, ... | | 著者 | Hou, C, Rohr, J, Tsodikov, O.V. | | 登録日 | 2016-05-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
5JW2
 
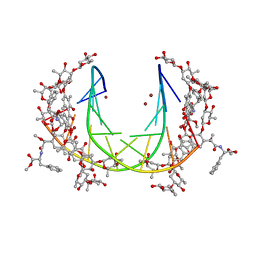 | | Crystal structure of mithramycin analogue MTM SA-Phe in complex with a 10-mer DNA AGGGATCCCT | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*AP*TP*CP*CP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Phe, ... | | 著者 | Hou, C, Rohr, J, Tsodikov, O.V. | | 登録日 | 2016-05-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
5JVT
 
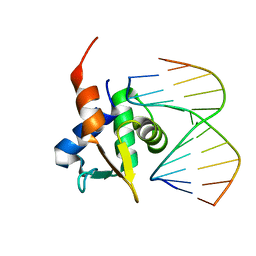 | |
7ZUD
 
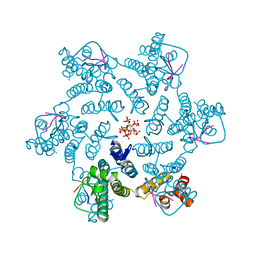 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | 分子名称: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | 著者 | Nicastro, G, Taylor, I.A. | | 登録日 | 2022-05-12 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
