2DH7
 
 | | Solution structure of the second RNA binding domain in Nucleolysin TIAR | | 分子名称: | Nucleolysin TIAR | | 著者 | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the second RNA binding domain in Nucleolysin TIAR
To be Published
|
|
2DHA
 
 | | Solution structure of the second RNA recognition motif in Hypothetical protein FLJ201171 | | 分子名称: | FLJ20171 protein | | 著者 | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the second RNA recognition motif in Hypothetical protein FLJ201171
To be Published
|
|
2DHG
 
 | | Solution structure of the C-terminal RNA recognition motif in tRNA selenocysteine associated protein | | 分子名称: | tRNA selenocysteine associated protein (SECP43) | | 著者 | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal RNA recognition motif in tRNA selenocysteine associated protein
To be Published
|
|
2RS2
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | 分子名称: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | 著者 | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | 登録日 | 2011-06-27 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
3AB5
 
 | |
2MST
 
 | | MUSASHI1 RBD2, NMR | | 分子名称: | PROTEIN (MUSASHI1) | | 著者 | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | 登録日 | 1999-05-19 | | 公開日 | 2000-05-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
2MSS
 
 | | MUSASHI1 RBD2, NMR | | 分子名称: | PROTEIN (MUSASHI1) | | 著者 | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | 登録日 | 1999-05-19 | | 公開日 | 2000-05-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
5YQ0
 
 | | Crystal structure of secreted protein CofJ from ETEC. | | 分子名称: | CALCIUM ION, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2018-07-25 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YPZ
 
 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | 分子名称: | CofB, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.521 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | 分子名称: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | 著者 | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | 分子名称: | RNA-binding protein Musashi homolog 1 | | 著者 | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
3WCQ
 
 | | Crystal structure analysis of Cyanidioschyzon melorae ferredoxin D58N mutant | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | 著者 | Ueno, Y, Matsumoto, T, Yamano, A, Imai, T, Morimoto, Y. | | 登録日 | 2013-05-31 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Increasing the electron-transfer ability of Cyanidioschyzon merolae ferredoxin by a one-point mutation - A high resolution and Fe-SAD phasing crystal structure analysis of the Asp58Asn mutant
Biochem.Biophys.Res.Commun., 436, 2013
|
|
8X6R
 
 | | KRasG12C in complex with inhibitor | | 分子名称: | 1-[7-[6-ethenyl-8-ethoxy-7-(5-methyl-1~{H}-indazol-4-yl)-2-(1-methylpiperidin-4-yl)oxy-quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Amano, Y, Tateishi, Y. | | 登録日 | 2023-11-21 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of ASP6918, a KRAS G12C inhibitor: Synthesis and structure-activity relationships of 1-{2,7-diazaspiro[3.5]non-2-yl}prop-2-en-1-one derivatives as covalent inhibitors with good potency and oral activity for the treatment of solid tumors.
Bioorg.Med.Chem., 98, 2023
|
|
7YCE
 
 | | KRas G12C in complex with Compound 7b | | 分子名称: | 1-[7-[6-chloranyl-2-(1-ethylpiperidin-4-yl)oxy-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Amano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
7YCC
 
 | | KRas G12C in complex with Compound 5c | | 分子名称: | 1-[7-[6-chloranyl-8-fluoranyl-7-(5-methyl-1~{H}-indazol-4-yl)-2-[(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Amano, Y. | | 登録日 | 2022-07-01 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery and biological evaluation of 1-{2,7-diazaspiro[3.5]nonan-2-yl}prop-2-en-1-one derivatives as covalent inhibitors of KRAS G12C with favorable metabolic stability and anti-tumor activity.
Bioorg.Med.Chem., 71, 2022
|
|
1UAW
 
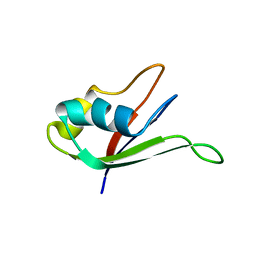 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | 分子名称: | mouse-musashi-1 | | 著者 | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | 登録日 | 2003-03-24 | | 公開日 | 2004-03-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
5AX6
 
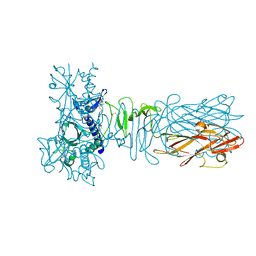 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | 分子名称: | ACETATE ION, CofB | | 著者 | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | 登録日 | 2015-07-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
2FDT
 
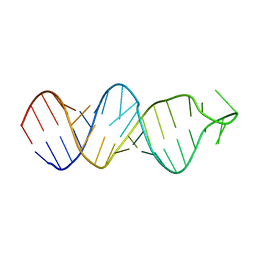 | | Solution structure of a conserved RNA hairpin of eel LINE UnaL2 | | 分子名称: | 36-mer | | 著者 | Nomura, Y, Baba, S, Nakazato, S, Sakamoto, T, Kajikawa, M, Okada, N, Kawai, G. | | 登録日 | 2005-12-14 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2
Nucleic Acids Res., 34, 2006
|
|
3WZY
 
 | |
3WZW
 
 | |
3WZV
 
 | |
3WZX
 
 | |
5SVW
 
 | |
5SVU
 
 | |
5SVG
 
 | |
