5CU1
 
 | |
4UQ3
 
 | | Crystal structure of HLA-A0201 in complex with an azobenzene- containing peptide | | 分子名称: | AZOBENZENE-CONTAINING PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | 著者 | Thong, S.Y, Yap, J.W, Lim, P.Y, Verhelst, S.H, Lescar, J, Meijers, R, Grotenbreg, G.M. | | 登録日 | 2014-06-19 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Bioorthogonal cleavage and exchange of major histocompatibility complex ligands by employing azobenzene-containing peptides.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4UQ2
 
 | | Crystal structure of HLA-A1101 in complex with an azobenzene- containing peptide | | 分子名称: | AZOBENZENE-CONTAINING PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | 著者 | Thong, S.Y, Yap, J.W, Lim, P.Y, Verhelst, S.H, Lescar, J, Meijers, R, Grotenbreg, G.M. | | 登録日 | 2014-06-19 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Bioorthogonal Cleavage and Exchange of Major Histocompatibility Complex Ligands by Employing Azobenzene-Containing Peptides.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2QKY
 
 | | complex structure of dipeptidyl peptidase IV and a oxadiazolyl ketone | | 分子名称: | 2-[(2-{(2S,4S)-2-[(R)-(5-tert-butyl-1,3,4-oxadiazol-2-yl)(hydroxy)methyl]-4-fluoropyrrolidin-1-yl}-2-oxoethyl)amino]-2-methylpropan-1-ol, Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) (Dipeptidyl peptidase 4 soluble form) (Dipeptidyl peptidase IV soluble form) | | 著者 | Kim, K.-H, Hong, S.Y, Koo, K.D, Lee, C.-S, Kim, G.T, Han, H.O. | | 登録日 | 2007-07-12 | | 公開日 | 2008-07-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Synthesis, SAR, and X-ray structure of novel potent DPPIV inhibitors: oxadiazolyl ketones.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2XU7
 
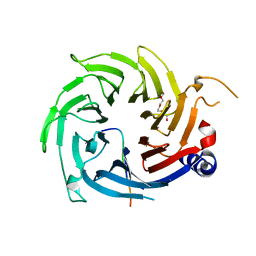 | | Structural basis for RbAp48 binding to FOG-1 | | 分子名称: | HISTONE-BINDING PROTEIN RBBP4, TETRAETHYLENE GLYCOL, ZINC FINGER PROTEIN ZFPM1 | | 著者 | Lejon, S, Thong, S.Y, Murthy, A, Blobel, G.A, Mackay, J.P, Murzina, N.V, Laue, E.D. | | 登録日 | 2010-10-15 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights Into Association of the Nurd Complex with Fog-1 from the Crystal Structure of an Rbap48-Fog- 1 Complex.
J.Biol.Chem., 286, 2011
|
|
1L3X
 
 | |
2KAE
 
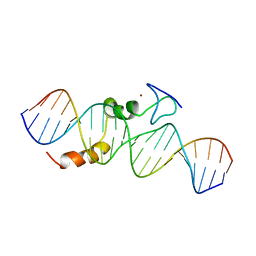 | | data-driven model of MED1:DNA complex | | 分子名称: | 5'-D(*DCP*DGP*DGP*DAP*DAP*DAP*DAP*DGP*DTP*DAP*DTP*DAP*DCP*DTP*DTP*DTP*DTP*DCP*DCP*DG)-3', GATA-type transcription factor, ZINC ION | | 著者 | Lowry, J.A, Gamsjaeger, R, Thong, S, Hung, W, Kwan, A.H, Broitman-Maduro, G, Matthews, J.M, Maduro, M, Mackay, J.P. | | 登録日 | 2008-11-04 | | 公開日 | 2009-01-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of MED-1 Reveals Unexpected Diversity in the Mechanism of DNA Recognition by GATA-type Zinc Finger Domains.
J.Biol.Chem., 284, 2009
|
|
2MPL
 
 | |
