6CI4
 
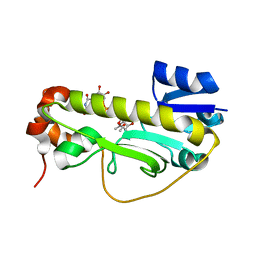 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis soaked with UDP-4-amino-4,6-dideoxy-L-AltNAc | | 分子名称: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-amino-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, formyltransferase PseJ | | 著者 | Harb, I, Reimer, J.M, Schmeing, T.M. | | 登録日 | 2018-02-23 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.824068 Å) | | 主引用文献 | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6EDK
 
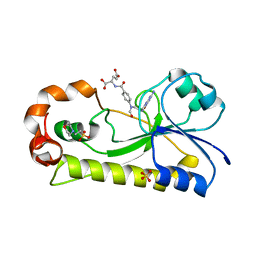 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis with N10-formyltetrahydrofolate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Formyltransferase PseJ, N-{4-[{[(6S)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}(formyl)amino]benzoyl}-L-glutamic acid, ... | | 著者 | Reimer, J.M, Harb, I, Schmeing, T.M. | | 登録日 | 2018-08-09 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6CI5
 
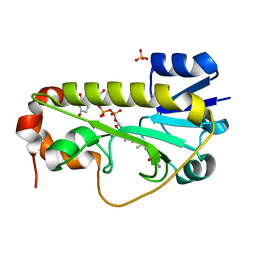 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis in complex with UDP-4,6-dideoxy-4-formamido-L-AltNAc and tetrahydrofolate | | 分子名称: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-(formylamino)-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-[4-({[(6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | 著者 | Reimer, J.M, Harb, I, Schmeing, T.M. | | 登録日 | 2018-02-23 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.00003052 Å) | | 主引用文献 | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6CI2
 
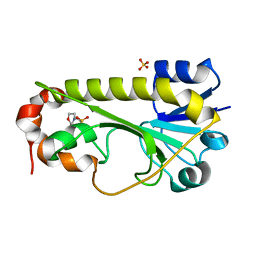 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, formyltransferase PseJ | | 著者 | Reimer, J.M, Jiang, J, Harb, I, Schmeing, T.M. | | 登録日 | 2018-02-23 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6MFY
 
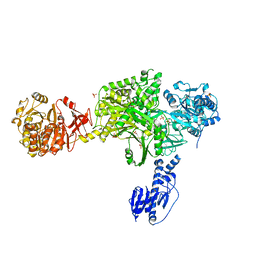 | | Crystal structure of a 5-domain construct of LgrA in the substrate donation state | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A, PHOSPHATE ION | | 著者 | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFZ
 
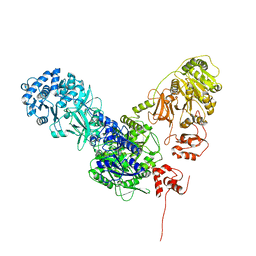 | | Crystal structure of dimodular LgrA in a condensation state | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A | | 著者 | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MG0
 
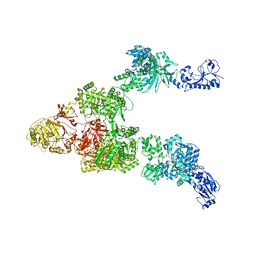 | | Crystal structure of a 5-domain construct of LgrA in the thiolation state | | 分子名称: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, Linear gramicidin synthase subunit A | | 著者 | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (6 Å) | | 主引用文献 | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFW
 
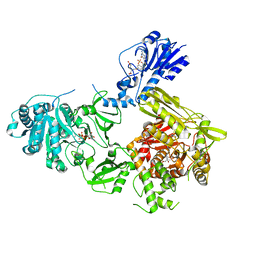 | | Crystal structure of a 4-domain construct of LgrA in the substrate donation state | | 分子名称: | (2~{R})-~{N}-[3-[2-[[(2~{S})-2-formamido-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]-3,3-dimethyl-2-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-butanamide, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, ... | | 著者 | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFX
 
 | | Crystal structure of a 4-domain construct of a mutant of LgrA in the substrate donation state | | 分子名称: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | 著者 | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
