6YL5
 
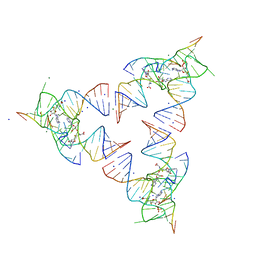 | | Crystal structure of the SAM-SAH riboswitch with SAH | | 分子名称: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YMJ
 
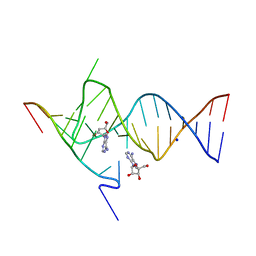 | | Crystal structure of the SAM-SAH riboswitch with adenosine. | | 分子名称: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE, Chains: A,C,F,I,M,O, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMK
 
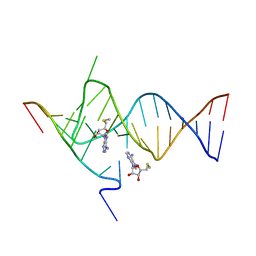 | | Crystal structure of the SAM-SAH riboswitch with AMP | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMI
 
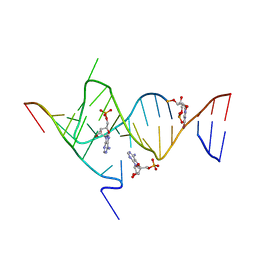 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | 分子名称: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-08 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
7LDE
 
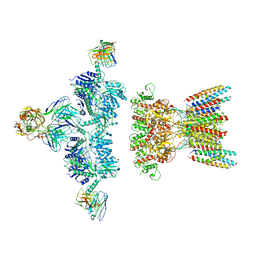 | | native AMPA receptor | | 分子名称: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDD
 
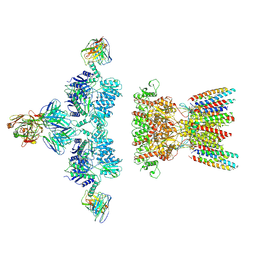 | | native AMPA receptor | | 分子名称: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LEP
 
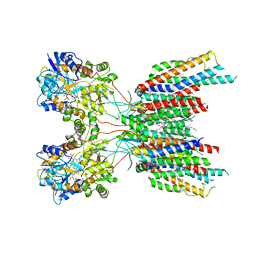 | | The composite LBD-TMD structure combined from all hippocampal AMPAR subtypes at 3.25 Angstrom resolution | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, DECANE, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-14 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
3LFU
 
 | |
6YLB
 
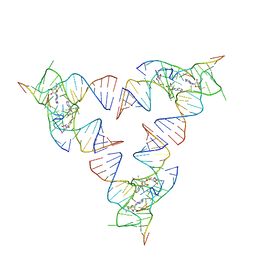 | | Crystal structure of the SAM-SAH riboswitch with SAM | | 分子名称: | Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, S-ADENOSYLMETHIONINE | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YML
 
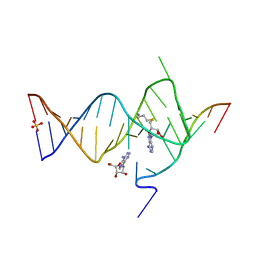 | |
6YMM
 
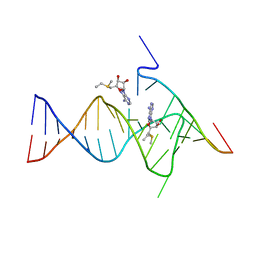 | |
4NL8
 
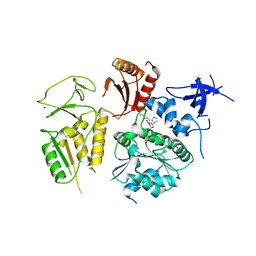 | | PriA Helicase Bound to SSB C-terminal Tail Peptide | | 分子名称: | Primosome assembly protein PriA, Single-stranded DNA-binding protein, ZINC ION | | 著者 | Bhattacharyya, B, George, N.P, Thurmes, T.M, Keck, J.L. | | 登録日 | 2013-11-13 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (4.08 Å) | | 主引用文献 | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NL4
 
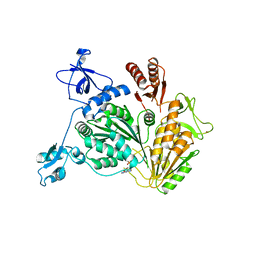 | |
6BHX
 
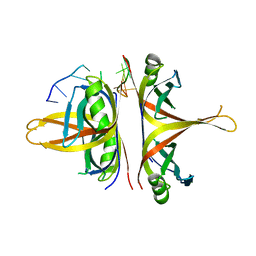 | | B. subtilis SsbA with DNA | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein A | | 著者 | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | 登録日 | 2017-10-31 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.936 Å) | | 主引用文献 | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
6BHW
 
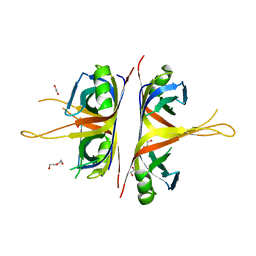 | | B. subtilis SsbA | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Single-stranded DNA-binding protein A | | 著者 | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | 登録日 | 2017-10-31 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.208 Å) | | 主引用文献 | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
5UK5
 
 | |
2GTE
 
 | | Drosophila OBP LUSH bound to attractant pheromone 11-cis-vaccenyl acetate | | 分子名称: | (Z)-OCTADEC-11-ENYL ACETATE, General odorant-binding protein lush, PHOSPHATE ION | | 著者 | Laughlin, J.D, Ha, T, Smith, D.P, Jones, D.N.M. | | 登録日 | 2006-04-27 | | 公開日 | 2007-06-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein
Cell(Cambridge,Mass.), 133, 2008
|
|
6L3G
 
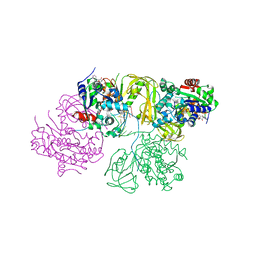 | | Structural Basis for DNA Unwinding at Forked dsDNA by two coordinating Pif1 helicases | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*TP*TP*TP*T)-3'), ... | | 著者 | Su, N, Bharath, S.R, Song, H. | | 登録日 | 2019-10-10 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for DNA unwinding at forked dsDNA by two coordinating Pif1 helicases.
Nat Commun, 10, 2019
|
|
