5ZH8
 
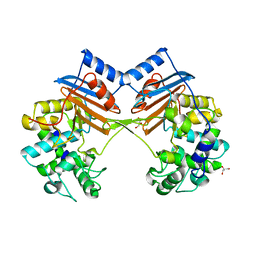 | | Crystal Structure of FmtA from Staphylococcus aureus at 2.58 A | | 分子名称: | GLYCEROL, Protein FmtA | | 著者 | Dalal, V, Kumar, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2018-03-12 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Repurposing an Ancient Protein Core Structure: Structural Studies on FmtA, a Novel Esterase of Staphylococcus aureus.
J.Mol.Biol., 431, 2019
|
|
1YT4
 
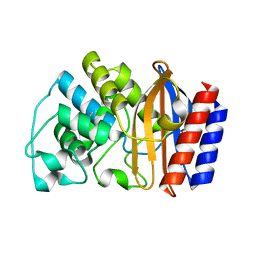 | | Crystal structure of TEM-76 beta-lactamase at 1.4 Angstrom resolution | | 分子名称: | Beta-lactamase TEM | | 著者 | Thomas, V.L, Golemi-Kotra, D, Kim, C, Vakulenko, S.B, Mobashery, S, Shoichet, B.K. | | 登録日 | 2005-02-09 | | 公開日 | 2005-07-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Consequences of the Inhibitor-Resistant Ser130Gly Substitution in TEM beta-Lactamase.
Biochemistry, 44, 2005
|
|
4Y0O
 
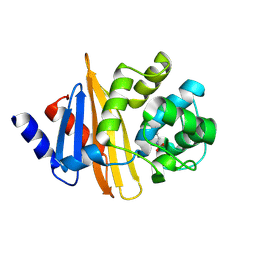 | | Crystal structure of OXA-58, a carbapenem hydrolyzing Class D beta-lactamase from Acinetobacter baumanii. | | 分子名称: | Beta-lactamase | | 著者 | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2015-02-06 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
4Y0U
 
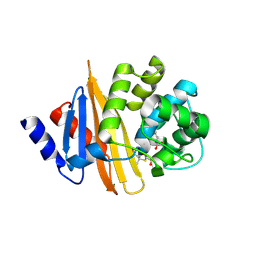 | | Crystal Structure of 6Alpha-Hydroxymethylpenicillanate Complexed with OXA-58, a Carbapenem hydrolyzing Class D betalactamase from Acinetobacter baumanii. | | 分子名称: | 2-(1-CARBOXY-2-HYDROXY-ETHYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Beta-lactamase | | 著者 | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2015-02-06 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
4Y0T
 
 | | Crystal structure of apo form of OXA-58, a Carbapenem hydrolyzing Class D beta-lactamase from Acinetobacter baumanii (P21, 4mol/ASU) | | 分子名称: | Beta-lactamase | | 著者 | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2015-02-06 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
4Z9Q
 
 | |
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | 分子名称: | pGolemi | | 著者 | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | 登録日 | 2008-08-03 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
6M8O
 
 | |
4IF4
 
 | |
4GVP
 
 | |
